Disease risk of missense mutations using structural inference from predicted function
- PMID: 20887259
- PMCID: PMC3095817
- DOI: 10.2174/138920310794109139
Disease risk of missense mutations using structural inference from predicted function
Abstract
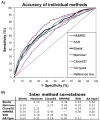
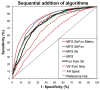
Advancements in sequencing techniques place personalized genomic medicine upon the horizon, bringing along the responsibility of clinicians to understand the likelihood for a mutation to cause disease, and of scientists to separate etiology from nonpathologic variability. Pathogenicity is discernable from patterns of interactions between a missense mutation, the surrounding protein structure, and intermolecular interactions. Physicochemical stability calculations are not accessible without structures, as is the case for the vast majority of human proteins, so diagnostic accuracy remains in infancy. To model the effects of missense mutations on functional stability without structure, we combine novel protein sequence analysis algorithms to discern spatial distributions of sequence, evolutionary, and physicochemical conservation, through a new approach to optimize component selection. Novel components include a combinatory substitution matrix and two heuristic algorithms that detect positions which confer structural support to interaction interfaces. The method reaches 0.91 AUC in ten-fold cross-validation to predict alteration of function for 6,392 in vitro mutations. For clinical utility we trained the method on 7,022 disease associated missense mutations within the Online Mendelian inheritance in man amongst a larger randomized set. In a blinded prospective test to delineate mutations unique to 186 patients with craniosynostosis from those in the 95 highly variant Coriell controls and 1000 age matched controls, we achieved roughly 1/3 sensitivity and perfect specificity. The component algorithms retained during machine learning constitute novel protein sequence analysis techniques to describe environments supporting neutrality or pathology of mutations. This approach to pathogenetics enables new insight into the mechanistic relationship of missense mutations to disease phenotypes in our patients.
Figures




Similar articles
-
Analyzing effects of naturally occurring missense mutations.Comput Math Methods Med. 2012;2012:805827. doi: 10.1155/2012/805827. Epub 2012 Apr 22. Comput Math Methods Med. 2012. PMID: 22577471 Free PMC article.
-
Detailed computational study of p53 and p16: using evolutionary sequence analysis and disease-associated mutations to predict the functional consequences of allelic variants.Oncogene. 2003 Feb 27;22(8):1150-63. doi: 10.1038/sj.onc.1206101. Oncogene. 2003. PMID: 12606942
-
Sequence-only evolutionary and predicted structural features for the prediction of stability changes in protein mutants.BMC Bioinformatics. 2013;14 Suppl 2(Suppl 2):S6. doi: 10.1186/1471-2105-14-S2-S6. Epub 2013 Jan 21. BMC Bioinformatics. 2013. PMID: 23369338 Free PMC article.
-
Accurate proteome-wide missense variant effect prediction with AlphaMissense.Science. 2023 Sep 22;381(6664):eadg7492. doi: 10.1126/science.adg7492. Epub 2023 Sep 22. Science. 2023. PMID: 37733863
-
Computational approaches for predicting mutant protein stability.J Comput Aided Mol Des. 2016 May;30(5):401-12. doi: 10.1007/s10822-016-9914-3. Epub 2016 May 9. J Comput Aided Mol Des. 2016. PMID: 27160393 Review.
Cited by
-
Exome sequencing identifies a recurrent de novo ZSWIM6 mutation associated with acromelic frontonasal dysostosis.Am J Hum Genet. 2014 Aug 7;95(2):235-40. doi: 10.1016/j.ajhg.2014.07.008. Am J Hum Genet. 2014. PMID: 25105228 Free PMC article.
References
-
- Wang Z, Moult J. SNPs, Protein Structure, and Disease. Hum Mutat. 2001;17(4):263–270. - PubMed
-
- Guerois R, Nielsen JE, Serrano L. Predicting changes in the stability of proteins and protein complexes: A study of more than 1000 mutations. J Mol Biol. 2002;320(2):369–387. - PubMed
-
- Yue P, Li Z, Moult J. Loss of protein structure stability as a major causative factor in monogenic disease. J Mol Biol. 2005;353(2):459–473. - PubMed
-
- Pakula AA, Sauer RT. Genetic analysis of protein stability and function. Annu Rev Genet. 1989;23:289–310. - PubMed
-
- Allali-Hassani A, Wasney GA, Chau I, Hong BS, Senisterra G, Loppnau P, Shi Z, Moult J, Edwards AM, Arrowsmith CH, Park HW, Schapira M, Vedadi M. A survey of proteins encoded by non-synonymous single nucleotide polymorphisms reveals a significant fraction with altered stability and activity. Biochem J. 2009;424(1):15–26. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

