Predicting the melting point of human C-type lysozyme mutants
- PMID: 20887260
- PMCID: PMC4667962
- DOI: 10.2174/138920310794109210
Predicting the melting point of human C-type lysozyme mutants
Abstract
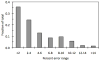
A complete understanding of the relationships between protein structure and stability remains an open problem. Much of our insight comes from laborious experimental analyses that perturb structure via directed mutation. The glycolytic enzyme lysozyme is among the most well characterized proteins under this paradigm, due to its abundance and ease of manipulation. To speed up such analyses, efficient computational models that can accurately predict mutation effects are needed. We employ a minimal Distance Constraint Model (mDCM) to predict the stability of a series of lysozyme mutants (specifically, human wild-type C-type lysozyme and 14 point mutations). With three phenomenological parameters that characterize microscopic interactions, the mDCM parameters are determined by obtaining the least squares error between predicted and experimental heat capacity curves. The mutants are chemically and structurally diverse, but have been experimentally characterized under nearly identical thermodynamic conditions (pH, ionic strength, etc.). The parameters found from best fits to heat capacity curves for one or more lysozyme structures are subsequently used to predict the heat capacity on the remaining. We simulate a typical experimental situation, where prediction of relative stabilities in an untested mutated structure is based on known results as they accumulate. From the statistical significance of these simulations, we establish that the mDCM is a viable predictor for relative stability of protein mutants. Remarkably, using parameters from any single fitting yields an average percent error of 4.3%. Across the dataset, the mDCM reproduces experimental trends sufficiently well (R = 0.64) to be of practical value to experimentalists when making decisions about which mutations to invest time and funds for characterization.
Figures
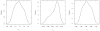




Similar articles
-
Hydrogen bonding and solvent structure in an antigen-antibody interface. Crystal structures and thermodynamic characterization of three Fv mutants complexed with lysozyme.Biochemistry. 1996 Dec 3;35(48):15494-503. doi: 10.1021/bi961709e. Biochemistry. 1996. PMID: 8952503
-
Stabilization of proteins by enhancement of inter-residue hydrophobic contacts: lessons of T4 lysozyme and barnase.J Biomol Struct Dyn. 2000 Dec;18(3):477-91. doi: 10.1080/07391102.2000.10506682. J Biomol Struct Dyn. 2000. PMID: 11149522
-
Contribution of hydrogen bonds to the conformational stability of human lysozyme: calorimetry and X-ray analysis of six Ser --> Ala mutants.Biochemistry. 1999 May 18;38(20):6623-9. doi: 10.1021/bi9901228. Biochemistry. 1999. PMID: 10350481
-
Contribution of hydrophobic residues to the stability of human lysozyme: calorimetric studies and X-ray structural analysis of the five isoleucine to valine mutants.J Mol Biol. 1995 Nov 17;254(1):62-76. doi: 10.1006/jmbi.1995.0599. J Mol Biol. 1995. PMID: 7473760
-
Applications of Protein Thermodynamic Database for Understanding Protein Mutant Stability and Designing Stable Mutants.Methods Mol Biol. 2016;1415:71-89. doi: 10.1007/978-1-4939-3572-7_4. Methods Mol Biol. 2016. PMID: 27115628
Cited by
-
Application of Rigidity Theory to the Thermostabilization of Lipase A from Bacillus subtilis.PLoS Comput Biol. 2016 Mar 22;12(3):e1004754. doi: 10.1371/journal.pcbi.1004754. eCollection 2016 Mar. PLoS Comput Biol. 2016. PMID: 27003415 Free PMC article.
-
Analyzing effects of naturally occurring missense mutations.Comput Math Methods Med. 2012;2012:805827. doi: 10.1155/2012/805827. Epub 2012 Apr 22. Comput Math Methods Med. 2012. PMID: 22577471 Free PMC article.
-
Calculating ensemble averaged descriptions of protein rigidity without sampling.PLoS One. 2012;7(2):e29176. doi: 10.1371/journal.pone.0029176. Epub 2012 Feb 22. PLoS One. 2012. PMID: 22383947 Free PMC article.
-
Mutations in Antibody Fragments Modulate Allosteric Response Via Hydrogen-Bond Network Fluctuations.Biophys J. 2016 May 10;110(9):1933-42. doi: 10.1016/j.bpj.2016.03.033. Biophys J. 2016. PMID: 27166802 Free PMC article.
-
Redistribution of flexibility in stabilizing antibody fragment mutants follows Le Châtelier's principle.PLoS One. 2014 Mar 26;9(3):e92870. doi: 10.1371/journal.pone.0092870. eCollection 2014. PLoS One. 2014. PMID: 24671209 Free PMC article.
References
-
- Huang LT, Saraboji K, Ho SY, Hwang SF, Ponnuswamy MN, Gromiha MM. Prediction of protein mutant stability using classification and regression tool. Biophys Chem. 2007;125(2–3):462–470. - PubMed
-
- Huang LT, Gromiha MM, Ho SY. Sequence analysis and rule development of predicting protein stability change upon mutation using decision tree model. J Mol Model. 2007;13(8):879–890. - PubMed
-
- Huang LT, Gromiha MM, Ho SY. iPTREE-STAB: interpretable decision tree based method for predicting protein stability changes upon mutations. Bioinformatics. 2007;23(10):1292–1293. - PubMed
-
- Cheng J, Randall A, Baldi P. Prediction of protein stability changes for single-site mutations using support vector machines. Proteins. 2006;62(4):1125–1132. - PubMed
-
- Capriotti E, Fariselli P, Calabrese R, Casadio R. Predicting protein stability changes from sequences using support vector machines. Bioinformatics. 2005;21(Suppl 2):ii54–58. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

