Analyses of subnanometer resolution cryo-EM density maps
- PMID: 20888467
- PMCID: PMC3107677
- DOI: 10.1016/S0076-6879(10)83001-0
Analyses of subnanometer resolution cryo-EM density maps
Abstract
Today, electron cryomicroscopy (cryo-EM) can routinely achieve subnanometer resolutions of complex macromolecular assemblies. From a density map, one can extract key structural and functional information using a variety of computational analysis tools. At subnanometer resolution, these tools make it possible to isolate individual subunits, identify secondary structures, and accurately fit atomic models. With several cryo-EM studies achieving resolutions beyond 5Å, computational modeling and feature recognition tools have been employed to construct backbone and atomic models of the protein components directly from a density map. In this chapter, we describe several common classes of computational tools that can be used to analyze and model subnanometer resolution reconstructions from cryo-EM. A general protocol for analyzing subnanometer resolution density maps is presented along with a full description of steps used in analyzing the 4.3Å resolution structure of Mm-cpn.
Copyright © 2010 Elsevier Inc. All rights reserved.
Figures

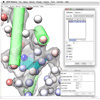



References
-
- Abeysinghe S, Ju T, Baker ML, Chiu W. Shape modeling and matching in identifying 3D protein structures. Comput. Aided Des. 2008;40:708–720.
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. - PubMed
-
- Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. - PubMed
-
- Baker ML, Jiang W, Bowman BR, Zhou ZH, Quiocho FA, Rixon FJ, Chiu W. Architecture of the herpes simplex virus major capsid protein derived from structural bioinformatics. J. Mol. Biol. 2003;331:447–456. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

