Classification of structural heterogeneity by maximum-likelihood methods
- PMID: 20888966
- PMCID: PMC3080752
- DOI: 10.1016/S0076-6879(10)82012-9
Classification of structural heterogeneity by maximum-likelihood methods
Abstract
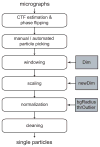
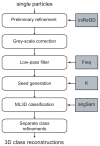
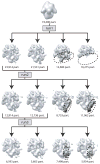
With the advent of computationally feasible approaches to maximum-likelihood (ML) image processing for cryo-electron microscopy, these methods have proven particularly useful in the classification of structurally heterogeneous single-particle data. A growing number of experimental studies have applied these algorithms to study macromolecular complexes with a wide range of structural variability, including nonstoichiometric complex formation, large conformational changes, and combinations of both. This chapter aims to share the practical experience that has been gained from the application of these novel approaches. Current insights on how to prepare the data and how to perform two- or three-dimensional classifications are discussed together with the aspects related to high-performance computing. Thereby, this chapter will hopefully be of practical use for those microscopists wishing to apply ML methods in their own investigations.
Copyright © 2010 Elsevier Inc. All rights reserved.
Figures



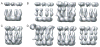

Similar articles
-
Processing of Structurally Heterogeneous Cryo-EM Data in RELION.Methods Enzymol. 2016;579:125-57. doi: 10.1016/bs.mie.2016.04.012. Epub 2016 May 31. Methods Enzymol. 2016. PMID: 27572726 Review.
-
SubspaceEM: A fast maximum-a-posteriori algorithm for cryo-EM single particle reconstruction.J Struct Biol. 2015 May;190(2):200-14. doi: 10.1016/j.jsb.2015.03.009. Epub 2015 Mar 31. J Struct Biol. 2015. PMID: 25839831 Free PMC article.
-
Likelihood-based structural analysis of electron microscopy images.Curr Opin Struct Biol. 2018 Apr;49:162-168. doi: 10.1016/j.sbi.2018.03.004. Epub 2018 Mar 24. Curr Opin Struct Biol. 2018. PMID: 29579548 Review.
-
Structural Study of Heterogeneous Biological Samples by Cryoelectron Microscopy and Image Processing.Biomed Res Int. 2017;2017:1032432. doi: 10.1155/2017/1032432. Epub 2017 Jan 15. Biomed Res Int. 2017. PMID: 28191458 Free PMC article. Review.
-
Particle migration analysis in iterative classification of cryo-EM single-particle data.J Struct Biol. 2014 Dec;188(3):267-73. doi: 10.1016/j.jsb.2014.10.006. Epub 2014 Oct 30. J Struct Biol. 2014. PMID: 25449317 Free PMC article.
Cited by
-
Semi-automated selection of cryo-EM particles in RELION-1.3.J Struct Biol. 2015 Feb;189(2):114-22. doi: 10.1016/j.jsb.2014.11.010. Epub 2014 Dec 6. J Struct Biol. 2015. PMID: 25486611 Free PMC article.
-
Current limits of structural biology: The transient interaction between cytochrome c 6 and photosystem I.Curr Res Struct Biol. 2020 Aug 26;2:171-179. doi: 10.1016/j.crstbi.2020.08.003. eCollection 2020. Curr Res Struct Biol. 2020. PMID: 34235477 Free PMC article.
-
Viral infection modulation and neutralization by camelid nanobodies.Proc Natl Acad Sci U S A. 2013 Apr 9;110(15):E1371-9. doi: 10.1073/pnas.1301336110. Epub 2013 Mar 25. Proc Natl Acad Sci U S A. 2013. PMID: 23530214 Free PMC article.
-
Dynamics in cryo EM reconstructions visualized with maximum-likelihood derived variance maps.J Struct Biol. 2013 Mar;181(3):195-206. doi: 10.1016/j.jsb.2012.11.005. Epub 2012 Dec 12. J Struct Biol. 2013. PMID: 23246781 Free PMC article.
-
A Representation Theory Perspective on Simultaneous Alignment and Classification.Appl Comput Harmon Anal. 2020 Nov;49(3):1001-1024. doi: 10.1016/j.acha.2019.05.005. Epub 2019 Jun 5. Appl Comput Harmon Anal. 2020. PMID: 39144545 Free PMC article.
References
-
- Al-Amoudi A, Diez DC, Betts MJ, Frangakis AS. The molecular architecture of cadherins in native epidermal desmosomes. Nature. 2007;450:832–837. - PubMed
-
- Bilbao-Castro J, Marabini R, Sorzano C, Garcia I, Carazo J, Fernandez J. Exploiting desktop supercomputing for three-dimensional electron microscopy reconstructions using ART with blobs. J Struct Biol. 2009;165:19–26. - PubMed
-
- Cheng K, Ivanova N, Scheres SH, Pavlov MY, Carazo JM, Hebert H, Ehrenberg M, Lindahl M. tmRNASmpB complex mimics native aminoacyl-tRNAs in the a site of stalled ribosomes. J Struct Biol. 2010;169:342–348. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

