FliZ regulates expression of the Salmonella pathogenicity island 1 invasion locus by controlling HilD protein activity in Salmonella enterica serovar typhimurium
- PMID: 20889744
- PMCID: PMC2981222
- DOI: 10.1128/JB.00635-10
FliZ regulates expression of the Salmonella pathogenicity island 1 invasion locus by controlling HilD protein activity in Salmonella enterica serovar typhimurium
Abstract
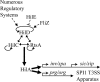
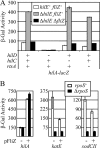
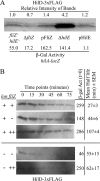
A prerequisite for Salmonella enterica to cause both intestinal and systemic disease is the direct injection of effector proteins into host intestinal epithelial cells via a type three secretion system (T3SS); the T3SS genes are carried on Salmonella pathogenicity island 1 (SPI1). These effector proteins induce inflammatory diarrhea and bacterial invasion. Expression of the SPI1 T3SS is tightly regulated in response to environmental signals through a variety of global regulatory systems. We have previously shown that three AraC-like regulators, HilD, HilC, and RtsA, act in a complex feed-forward regulatory loop to control the expression of the hilA gene, which encodes the direct regulator of the SPI1 structural genes. In this work, we characterize a major positive regulator of this system, the flagellar protein FliZ. Through genetic and biochemical analyses, we show that FliZ posttranslationally controls HilD to positively regulate hilA expression. This mechanism is independent of other flagellar components and is not mediated through the negative regulator HilE or through FliZ-mediated RpoS regulation. We demonstrate that FliZ controls HilD protein activity and not stability. FliZ regulates HilD in the absence of Lon protease, previously shown to degrade HilD. Indeed, it appears that FliZ, rather than HilD, is the most relevant target of Lon as it relates to SPI1 expression. Mutants lacking FliZ are significantly attenuated in their ability to colonize the intestine but are unaffected during systemic infection. The intestinal attenuation is partially dependent on SPI1, but FliZ has additional pleiotropic effects.
Figures



Similar articles
-
HilE Regulates HilD by Blocking DNA Binding in Salmonella enterica Serovar Typhimurium.J Bacteriol. 2018 Mar 26;200(8):e00750-17. doi: 10.1128/JB.00750-17. Print 2018 Apr 15. J Bacteriol. 2018. PMID: 29378886 Free PMC article.
-
HilD, HilC and RtsA constitute a feed forward loop that controls expression of the SPI1 type three secretion system regulator hilA in Salmonella enterica serovar Typhimurium.Mol Microbiol. 2005 Aug;57(3):691-705. doi: 10.1111/j.1365-2958.2005.04737.x. Mol Microbiol. 2005. PMID: 16045614
-
HilD, HilC, and RtsA Form Homodimers and Heterodimers To Regulate Expression of the Salmonella Pathogenicity Island I Type III Secretion System.J Bacteriol. 2020 Apr 9;202(9):e00012-20. doi: 10.1128/JB.00012-20. Print 2020 Apr 9. J Bacteriol. 2020. PMID: 32041797 Free PMC article.
-
Adaptation to the host environment: regulation of the SPI1 type III secretion system in Salmonella enterica serovar Typhimurium.Curr Opin Microbiol. 2007 Feb;10(1):24-9. doi: 10.1016/j.mib.2006.12.002. Epub 2007 Jan 5. Curr Opin Microbiol. 2007. PMID: 17208038 Review.
-
Salmonella invasion gene regulation: a story of environmental awareness.J Microbiol. 2005 Feb;43 Spec No:110-7. J Microbiol. 2005. PMID: 15765064 Review.
Cited by
-
YeiE Regulates Motility and Gut Colonization in Salmonella enterica Serotype Typhimurium.mBio. 2021 Jun 29;12(3):e0368020. doi: 10.1128/mBio.03680-20. Epub 2021 Jun 8. mBio. 2021. PMID: 34098734 Free PMC article.
-
Salmonella Pathogenicity Island 1 (SPI-1) and Its Complex Regulatory Network.Front Cell Infect Microbiol. 2019 Jul 31;9:270. doi: 10.3389/fcimb.2019.00270. eCollection 2019. Front Cell Infect Microbiol. 2019. PMID: 31428589 Free PMC article. Review.
-
The Small RNA PinT Contributes to PhoP-Mediated Regulation of the Salmonella Pathogenicity Island 1 Type III Secretion System in Salmonella enterica Serovar Typhimurium.J Bacteriol. 2019 Sep 6;201(19):e00312-19. doi: 10.1128/JB.00312-19. Print 2019 Oct 1. J Bacteriol. 2019. PMID: 31262841 Free PMC article.
-
Experimental Evolution of Anticipatory Regulation in Escherichia coli.Front Microbiol. 2022 Jan 11;12:796228. doi: 10.3389/fmicb.2021.796228. eCollection 2021. Front Microbiol. 2022. PMID: 35087497 Free PMC article.
-
The motility regulator flhDC drives intracellular accumulation and tumor colonization of Salmonella.J Immunother Cancer. 2019 Feb 12;7(1):44. doi: 10.1186/s40425-018-0490-z. J Immunother Cancer. 2019. PMID: 30755273 Free PMC article.
References
-
- Altier, C. 2005. Genetic and environmental control of Salmonella invasion. J. Microbiol. 43:85-92. - PubMed
-
- Bajaj, V., C. Hwang, and C. A. Lee. 1995. HilA is a novel OmpR/ToxR family member that activates the expression of Salmonella typhimurium invasion genes. Mol. Microbiol. 18:715-727. - PubMed
-
- Bumann, D. 2002. Examination of Salmonella gene expression in an infected mammalian host using the green fluorescent protein and two-colour flow cytometry. Mol. Microbiol. 43:1269-1283. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

