Evaluation of virulence factor profiling in the characterization of veterinary Escherichia coli isolates
- PMID: 20889790
- PMCID: PMC2976202
- DOI: 10.1128/AEM.00726-10
Evaluation of virulence factor profiling in the characterization of veterinary Escherichia coli isolates
Abstract
Escherichia coli has been used as an indicator organism for fecal contamination of water and other environments and is often a commensal organism in healthy animals, yet a number of strains can cause disease in young or immunocompromised animals. In this study, 281 E. coli isolates from bovine, porcine, chicken, canine, equine, feline, and other veterinary sources were analyzed by BOXA1R PCR and by virulence factor profiling of 35 factors to determine whether they had utility in identifying the animal source of the isolates. The results of BOXA1R PCR analysis demonstrated a high degree of diversity; less than half of the isolates fell into one of 27 clusters with at least three isolates (based on 90% similarity). Nearly 60% of these clusters contained isolates from more than one animal source. Conversely, the results of virulence factor profiling demonstrated clustering by animal source. Three clusters, named Bovine, Chicken, and Porcine, based on discriminant components analysis, were represented by 90% or more of the respective isolates. A fourth group, termed Companion, was the most diverse, containing at least 84% of isolates from canine, feline, equine, and other animal sources. Based on these results, it appears that virulence factor profiling may have utility, helping identify the likely animal host species sources of certain E. coli isolates.
Figures
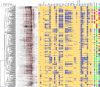
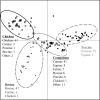
 ), Feline (
), Feline ( ), Equine (〉), and Other (
), Equine (〉), and Other ( ). Four major clusters were identified, delineated by ovals on the figure, and defined by the predominant source (Bovine, Chicken, Swine, and Companion). Adjacent to the ovals are the number of isolates from different animal species present in the respective clusters.
). Four major clusters were identified, delineated by ovals on the figure, and defined by the predominant source (Bovine, Chicken, Swine, and Companion). Adjacent to the ovals are the number of isolates from different animal species present in the respective clusters.Similar articles
-
Comparative analysis of virulence genes, genetic diversity, and phylogeny of commensal and enterotoxigenic Escherichia coli isolates from weaned pigs.Appl Environ Microbiol. 2007 Jan;73(1):83-91. doi: 10.1128/AEM.00990-06. Epub 2006 Oct 20. Appl Environ Microbiol. 2007. PMID: 17056683 Free PMC article.
-
Clustered, regularly interspaced short palindromic repeat (CRISPR) diversity and virulence factor distribution in avian Escherichia coli.Res Microbiol. 2017 Feb-Mar;168(2):147-156. doi: 10.1016/j.resmic.2016.10.002. Epub 2016 Oct 24. Res Microbiol. 2017. PMID: 27789334
-
Population structure and virulence content of avian pathogenic Escherichia coli isolated from outbreaks in Sri Lanka.Vet Microbiol. 2014 Jan 31;168(2-4):403-12. doi: 10.1016/j.vetmic.2013.11.028. Epub 2013 Dec 11. Vet Microbiol. 2014. PMID: 24388626
-
Genetic diversity and virulence profiles of Escherichia coli isolates causing spontaneous bacterial peritonitis and bacteremia in patients with cirrhosis.J Clin Microbiol. 2010 Aug;48(8):2709-14. doi: 10.1128/JCM.00516-10. Epub 2010 Jun 2. J Clin Microbiol. 2010. PMID: 20519468 Free PMC article.
-
Multidrug-resistant extraintestinal pathogenic Escherichia coli of sequence type ST131 in animals and foods.Vet Microbiol. 2011 Nov 21;153(1-2):99-108. doi: 10.1016/j.vetmic.2011.05.007. Epub 2011 May 13. Vet Microbiol. 2011. PMID: 21658865 Review.
Cited by
-
Evaluation of Incompatibility Group I1 (IncI1) Plasmid-Containing Salmonella enterica and Assessment of the Plasmids in Bacteriocin Production and Biofilm Development.Front Vet Sci. 2019 Sep 6;6:298. doi: 10.3389/fvets.2019.00298. eCollection 2019. Front Vet Sci. 2019. PMID: 31552285 Free PMC article.
-
Impact of plasmids, including those encodingVirB4/D4 type IV secretion systems, on Salmonella enterica serovar Heidelberg virulence in macrophages and epithelial cells.PLoS One. 2013 Oct 3;8(10):e77866. doi: 10.1371/journal.pone.0077866. eCollection 2013. PLoS One. 2013. PMID: 24098597 Free PMC article.
References
-
- Aiello, S. E., and A. Mays (ed.). 1998. The Merck veterinary manual, 8th ed. Merck and Company, Whitehouse Station, NJ.
-
- Auld, H., D. MacIver, and J. Klaassen. 2004. Heavy rainfall and waterborne disease outbreaks: the Walkerton example. J. Toxicol. Environ. Health A 67:1879-1887. - PubMed
-
- Barnes, H. J., J. P. Vaillancourt, and W. B. Gross. 2003. Colibacillosis, p. 631-656. In Y. M. Saif, H. J. Barnes, J. R. Glisson, A. M. Fadly, L. R. McDougald, and D. E. Swayne (ed.), Diseases of poultry, 11th ed. Iowa State University Press, Ames, IA.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

