Integration of a Notch-dependent mesenchymal gene program and Bmp2-driven cell invasiveness regulates murine cardiac valve formation
- PMID: 20890042
- PMCID: PMC2947227
- DOI: 10.1172/JCI42666
Integration of a Notch-dependent mesenchymal gene program and Bmp2-driven cell invasiveness regulates murine cardiac valve formation
Abstract
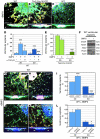
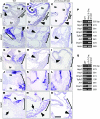
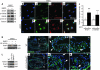
Cardiac valve formation is crucial for embryonic and adult heart function. Valve malformations constitute the most common congenital cardiac defect, but little is known about the molecular mechanisms regulating valve formation and homeostasis. Here, we show that endocardial Notch1 and myocardial Bmp2 signal integration establish a valve-forming field between 2 chamber developmental domains. Patterning occurs through the activation of endocardial epithelial-to-mesenchymal transition (EMT) exclusively in prospective valve territories. Mice with constitutive endocardial Notch1 activity ectopically express Hey1 and Heyl. They also display an activated mesenchymal gene program in ventricles and a partial (noninvasive) EMT in vitro that becomes invasive upon BMP2 treatment. Snail1, TGF-β2, or Notch1 inhibition reduces BMP2-induced ventricular transformation and invasion, whereas BMP2 treatment inhibits endothelial Gsk3β, stabilizing Snail1 and promoting invasiveness. Integration of Notch and Bmp2 signals is consistent with Notch1 signaling being attenuated after myocardial Bmp2 deletion. Notch1 activation in myocardium extends Hey1 expression to nonchamber myocardium, represses Bmp2, and impairs EMT. In contrast, Notch deletion abrogates endocardial Hey gene transcription and extends Bmp2 expression to the ventricular endocardium. This embryonic Notch1-Bmp2-Snail1 relationship may be relevant in adult valve disease, in which decreased NOTCH signaling causes valve mesenchyme cell formation, fibrosis, and calcification.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

