Functional imaging of hippocampal place cells at cellular resolution during virtual navigation
- PMID: 20890294
- PMCID: PMC2967725
- DOI: 10.1038/nn.2648
Functional imaging of hippocampal place cells at cellular resolution during virtual navigation
Abstract
Spatial navigation is often used as a behavioral task in studies of the neuronal circuits that underlie cognition, learning and memory in rodents. The combination of in vivo microscopy with genetically encoded indicators has provided an important new tool for studying neuronal circuits, but has been technically difficult to apply during navigation. Here we describe methods for imaging the activity of neurons in the CA1 region of the hippocampus with subcellular resolution in behaving mice. Neurons that expressed the genetically encoded calcium indicator GCaMP3 were imaged through a chronic hippocampal window. Head-restrained mice performed spatial behaviors in a setup combining a virtual reality system and a custom-built two-photon microscope. We optically identified populations of place cells and determined the correlation between the location of their place fields in the virtual environment and their anatomical location in the local circuit. The combination of virtual reality and high-resolution functional imaging should allow a new generation of studies to investigate neuronal circuit dynamics during behavior.
Figures


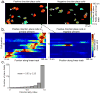
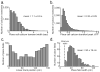
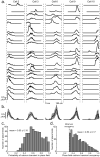
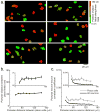

Comment in
-
Neuroscience in a virtual world.Nat Methods. 2010 Dec;7(12):948-9. doi: 10.1038/nmeth1210-948a. Nat Methods. 2010. PMID: 21158013 No abstract available.
References
-
- O'Keefe J, Dostrovsky J. The hippocampus as a spatial map. Preliminary evidence from unit activity in the freely-moving rat. Brain Res. 1971;34:171–175. - PubMed
-
- O'Keefe J. Hippocampal Neurophysiology in the Behaving Animal. In: Andersen P, editor. The hippocampus book. Oxford University Press; Oxford; New York: 2007. pp. 475–548.
-
- Leutgeb S, Leutgeb JK, Moser MB, Moser EI. Place cells, spatial maps and the population code for memory. Curr Opin Neurobiol. 2005;15:738–746. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

