The redox basis of epigenetic modifications: from mechanisms to functional consequences
- PMID: 20919933
- PMCID: PMC3118659
- DOI: 10.1089/ars.2010.3492
The redox basis of epigenetic modifications: from mechanisms to functional consequences
Abstract
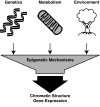
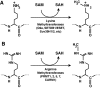
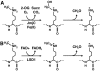
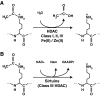
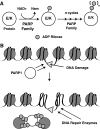
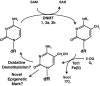
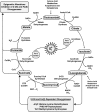
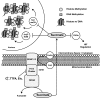
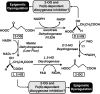
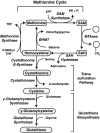
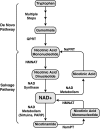
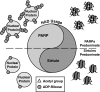
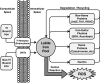
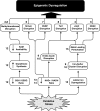
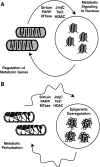
Epigenetic modifications represent mechanisms by which cells may effectively translate multiple signaling inputs into phenotypic outputs. Recent research is revealing that redox metabolism is an increasingly important determinant of epigenetic control that may have significant ramifications in both human health and disease. Numerous characterized epigenetic marks, including histone methylation, acetylation, and ADP-ribosylation, as well as DNA methylation, have direct linkages to central metabolism through critical redox intermediates such as NAD(+), S-adenosyl methionine, and 2-oxoglutarate. Fluctuations in these intermediates caused by both normal and pathologic stimuli may thus have direct effects on epigenetic signaling that lead to measurable changes in gene expression. In this comprehensive review, we present surveys of both metabolism-sensitive epigenetic enzymes and the metabolic processes that may play a role in their regulation. To close, we provide a series of clinically relevant illustrations of the communication between metabolism and epigenetics in the pathogenesis of cardiovascular disease, Alzheimer disease, cancer, and environmental toxicity. We anticipate that the regulatory mechanisms described herein will play an increasingly large role in our understanding of human health and disease as epigenetics research progresses.
Figures

References
-
- Ackrell BA. Cytopathies involving mitochondrial complex II. Mol Aspects Med. 2002;23:369–384. - PubMed
-
- Aghili M. Zahedi F. Rafiee E. Hydroxyglutaric aciduria and malignant brain tumor: a case report and literature review. J Neurooncol. 2009;91:233–236. - PubMed
-
- Ahringer J. NuRD and SIN3 histone deacetylase complexes in development. Trends Genet. 2000;16:351–356. - PubMed
-
- Allis CD. Berger SL. Cote J. Dent S. Jenuwien T. Kouzarides T. Pillus L. Reinberg D. Shi Y. Shiekhattar R, et al. New nomenclature for chromatin-modifying enzymes. Cell. 2007;131:633–636. - PubMed
-
- Althaus FR. Hofferer L. Kleczkowska HE. Malanga M. Naegeli H. Panzeter PL. Realini CA. Histone shuttling by poly ADP-ribosylation. Mol Cell Biochem. 1994;138:53–59. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

