Inferring the conservative causal core of gene regulatory networks
- PMID: 20920161
- PMCID: PMC2955605
- DOI: 10.1186/1752-0509-4-132
Inferring the conservative causal core of gene regulatory networks
Abstract
Background: Inferring gene regulatory networks from large-scale expression data is an important problem that received much attention in recent years. These networks have the potential to gain insights into causal molecular interactions of biological processes. Hence, from a methodological point of view, reliable estimation methods based on observational data are needed to approach this problem practically.
Results: In this paper, we introduce a novel gene regulatory network inference (GRNI) algorithm, called C3NET. We compare C3NET with four well known methods, ARACNE, CLR, MRNET and RN, conducting in-depth numerical ensemble simulations and demonstrate also for biological expression data from E. coli that C3NET performs consistently better than the best known GRNI methods in the literature. In addition, it has also a low computational complexity. Since C3NET is based on estimates of mutual information values in conjunction with a maximization step, our numerical investigations demonstrate that our inference algorithm exploits causal structural information in the data efficiently.
Conclusions: For systems biology to succeed in the long run, it is of crucial importance to establish methods that extract large-scale gene networks from high-throughput data that reflect the underlying causal interactions among genes or gene products. Our method can contribute to this endeavor by demonstrating that an inference algorithm with a neat design permits not only a more intuitive and possibly biological interpretation of its working mechanism but can also result in superior results.
Figures
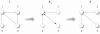





References
-
- Emmert-Streib F, Dehmer M, (Eds) Medical Biostatistics for Complex Diseases. Weinheim: Wiley-Blackwell; 2010.
-
- Alon U. An Introduction to Systems Biology: Design Principles of Biological Circuits. Boca Raton, FL: Chapman & Hall/CRC; 2006.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

