Comparative transcriptional profiling of the limbal epithelial crypt demonstrates its putative stem cell niche characteristics
- PMID: 20920242
- PMCID: PMC2997017
- DOI: 10.1186/1471-2164-11-526
Comparative transcriptional profiling of the limbal epithelial crypt demonstrates its putative stem cell niche characteristics
Abstract
Background: The Limbal epithelial crypt (LEC) is a solid cord of cells, approximately 120 microns long. It arises from the undersurface of interpalisade rete ridges of the limbal palisades of Vogt and extends deeper into the limbal stroma parallel or perpendicular to the palisade. There are up to 6 or 7 such LEC, variably distributed along the limbus in each human eye. Morphological and immunohistochemical studies on the limbal epithelial crypt (LEC) have demonstrated the presence of limbal stem cells in this region. The purpose of this microarray study was to characterise the transcriptional profile of the LEC and compare with other ocular surface epithelial regions to support our hypothesis that LEC preferentially harbours stem cells (SC).
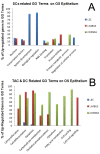
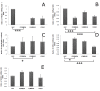
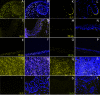
Results: LEC was found to be enriched for SC related Gene Ontology (GO) terms including those identified in quiescent adult SC, however similar to cornea, limbus had significant GO terms related to proliferating SC, transient amplifying cells (TAC) and differentiated cells (DC). LEC and limbus were metabolically dormant with low protein synthesis and downregulated cell cycling. Cornea had upregulated genes for cell cycling and self renewal such as FZD7, BTG1, CCNG, and STAT3 which were identified from other SC populations. Upregulated gene expression for growth factors, cytokines, WNT, Notch, TGF-Beta pathways involved in cell proliferation and differentiation were noted in cornea. LEC had highest number of expressed sequence tags (ESTs), downregulated and unknown genes, compared to other regions. Genes expressed in LEC such as CDH1, SERPINF1, LEF1, FRZB1, KRT19, SOD2, EGR1 are known to be involved in SC maintenance. Genes of interest, in LEC belonging to the category of cell adhesion molecules, WNT and Notch signalling pathway were validated with real-time PCR and immunofluorescence.
Conclusions: Our transcriptional profiling study identifies the LEC as a preferential site for limbal SC with some characteristics suggesting that it could function as a 'SC niche' supporting quiescent SC. It also strengthens the evidence for the presence of "transient cells" in the corneal epithelium. These cells are immediate progeny of SC with self-renewal capacity and could be responsible for maintaining epithelial turn over in normal healthy conditions of the ocular surface (OS). The limbus has mixed population of differentiated and undifferentiated cells.
Figures


References
-
- Schlotzer-Schrehardt U, Kruse FE. Identification and characterization of limbal stem cells. Exp Eye Res. 2005;81:247–264. - PubMed
-
- Kasper M. Patterns of cytokeratins and vimentin in guinea pig and mouse eye tissue: evidence for regional variations in intermediate filament expression in limbal epithelium. Acta Histochem. 1992;93:319–332. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

