The differential interaction of snRNPs with pre-mRNA reveals splicing kinetics in living cells
- PMID: 20921136
- PMCID: PMC2953428
- DOI: 10.1083/jcb.201004030
The differential interaction of snRNPs with pre-mRNA reveals splicing kinetics in living cells
Abstract
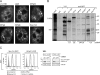
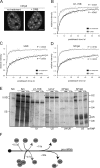
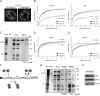
Precursor messenger RNA (pre-mRNA) splicing is catalyzed by the spliceosome, a large ribonucleoprotein (RNP) complex composed of five small nuclear RNP particles (snRNPs) and additional proteins. Using live cell imaging of GFP-tagged snRNP components expressed at endogenous levels, we examined how the spliceosome assembles in vivo. A comprehensive analysis of snRNP dynamics in the cell nucleus enabled us to determine snRNP diffusion throughout the nucleoplasm as well as the interaction rates of individual snRNPs with pre-mRNA. Core components of the spliceosome, U2 and U5 snRNPs, associated with pre-mRNA for 15-30 s, indicating that splicing is accomplished within this time period. Additionally, binding of U1 and U4/U6 snRNPs with pre-mRNA occurred within seconds, indicating that the interaction of individual snRNPs with pre-mRNA is distinct. These results are consistent with the predictions of the step-wise model of spliceosome assembly and provide an estimate on the rate of splicing in human cells.
Figures

References
-
- Behzadnia N., Golas M.M., Hartmuth K., Sander B., Kastner B., Deckert J., Dube P., Will C.L., Urlaub H., Stark H., Lührmann R. 2007. Composition and three-dimensional EM structure of double affinity-purified, human prespliceosomal A complexes. EMBO J. 26:1737–1748 10.1038/sj.emboj.7601631 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

