Young proteins experience more variable selection pressures than old proteins
- PMID: 20921233
- PMCID: PMC2963820
- DOI: 10.1101/gr.109595.110
Young proteins experience more variable selection pressures than old proteins
Abstract
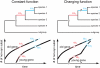
It is well known that young proteins tend to experience weaker purifying selection and evolve more quickly than old proteins. Here, we show that, in addition, young proteins tend to experience more variable selection pressures over time than old proteins. We demonstrate this pattern in three independent taxonomic groups: yeast, Drosophila, and mammals. The increased variability of selection pressures on young proteins is highly significant even after controlling for the fact that young proteins are typically shorter and experience weaker purifying selection than old proteins. The majority of our results are consistent with the hypothesis that the function of a young gene tends to change over time more readily than that of an old gene. At the same time, our results may be caused in part by young genes that serve constant functions over time, but nevertheless appear to evolve under changing selection pressures due to depletion of adaptive mutations. In either case, our results imply that the evolution of a protein-coding sequence is partly determined by its age and origin, and not only by the phenotypic properties of the encoded protein. We discuss, via specific examples, the consequences of these findings for understanding of the sources of evolutionary novelty.
Figures

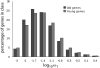
Similar articles
-
Relaxed purifying selection and possibly high rate of adaptation in primate lineage-specific genes.Genome Biol Evol. 2010 Jul 12;2:393-409. doi: 10.1093/gbe/evq019. Genome Biol Evol. 2010. PMID: 20624743 Free PMC article.
-
The efficiency of purifying selection in Mammals vs. Drosophila for metabolic genes.J Evol Biol. 2009 Oct;22(10):2118-24. doi: 10.1111/j.1420-9101.2009.01814.x. Epub 2009 Aug 20. J Evol Biol. 2009. PMID: 19694896
-
The relationship between the hierarchical position of proteins in the human signal transduction network and their rate of evolution.BMC Evol Biol. 2012 Sep 28;12:192. doi: 10.1186/1471-2148-12-192. BMC Evol Biol. 2012. PMID: 23020283 Free PMC article.
-
DNA sequence evolution: the sounds of silence.Philos Trans R Soc Lond B Biol Sci. 1995 Sep 29;349(1329):241-7. doi: 10.1098/rstb.1995.0108. Philos Trans R Soc Lond B Biol Sci. 1995. PMID: 8577834 Review.
-
Signatures of protein biophysics in coding sequence evolution.Curr Opin Struct Biol. 2010 Jun;20(3):385-9. doi: 10.1016/j.sbi.2010.03.004. Epub 2010 Apr 13. Curr Opin Struct Biol. 2010. PMID: 20395125 Free PMC article. Review.
Cited by
-
Are Antisense Proteins in Prokaryotes Functional?Front Mol Biosci. 2020 Aug 14;7:187. doi: 10.3389/fmolb.2020.00187. eCollection 2020. Front Mol Biosci. 2020. PMID: 32923454 Free PMC article.
-
What Signatures Dominantly Associate with Gene Age?Genome Biol Evol. 2016 Oct 13;8(10):3083-3089. doi: 10.1093/gbe/evw216. Genome Biol Evol. 2016. PMID: 27609935 Free PMC article.
-
Structure and age jointly influence rates of protein evolution.PLoS Comput Biol. 2012 May;8(5):e1002542. doi: 10.1371/journal.pcbi.1002542. Epub 2012 May 31. PLoS Comput Biol. 2012. PMID: 22693443 Free PMC article.
-
Genomic Complexity Places Less Restrictions on the Evolution of Young Coexpression Networks than Protein-Protein Interactions.Genome Biol Evol. 2016 Sep 3;8(8):2624-31. doi: 10.1093/gbe/evw198. Genome Biol Evol. 2016. PMID: 27521813 Free PMC article.
-
Factors That Affect the Rates of Adaptive and Nonadaptive Evolution at the Gene Level in Humans and Chimpanzees.Genome Biol Evol. 2022 Feb 4;14(2):evac028. doi: 10.1093/gbe/evac028. Genome Biol Evol. 2022. PMID: 35166775 Free PMC article.
References
-
- Alba MM, Castresana J 2005. Inverse relationship between evolutionary rate and age of mammalian genes. Mol Biol Evol 22: 598–606 - PubMed
-
- Barth AL, Dugas JC, Ngai J 1997. Noncoordinate expression of odorant receptor genes tightly linked in the zebrafish genome. Neuron 19: 359–369 - PubMed
-
- Buck L, Axel R 1991. A novel multigene family may encode odorant receptors: A molecular basis for odor recognition. Cell 65: 175–187 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
