Long-range tertiary interactions in single hammerhead ribozymes bias motional sampling toward catalytically active conformations
- PMID: 20921269
- PMCID: PMC2995402
- DOI: 10.1261/rna.1829110
Long-range tertiary interactions in single hammerhead ribozymes bias motional sampling toward catalytically active conformations
Abstract
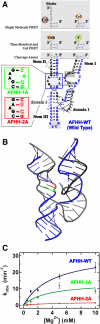
Enzymes generally are thought to derive their functional activity from conformational motions. The limited chemical variation in RNA suggests that such structural dynamics may play a particularly important role in RNA function. Minimal hammerhead ribozymes are known to cleave efficiently only in ∼ 10-fold higher than physiologic concentrations of Mg(2+) ions. Extended versions containing native loop-loop interactions, however, show greatly enhanced catalytic activity at physiologically relevant Mg(2+) concentrations, for reasons that are still ill-understood. Here, we use Mg(2+) titrations, activity assays, ensemble, and single molecule fluorescence resonance energy transfer (FRET) approaches, combined with molecular dynamics (MD) simulations, to ask what influence the spatially distant tertiary loop-loop interactions of an extended hammerhead ribozyme have on its structural dynamics. By comparing hammerhead variants with wild-type, partially disrupted, and fully disrupted loop-loop interaction sequences we find that the tertiary interactions lead to a dynamic motional sampling that increasingly populates catalytically active conformations. At the global level the wild-type tertiary interactions lead to more frequent, if transient, encounters of the loop-carrying stems, whereas at the local level they lead to an enrichment in favorable in-line attack angles at the cleavage site. These results invoke a linkage between RNA structural dynamics and function and suggest that loop-loop interactions in extended hammerhead ribozymes-and Mg(2+) ions that bind to minimal ribozymes-may generally allow more frequent access to a catalytically relevant conformation(s), rather than simply locking the ribozyme into a single active state.
Figures




Similar articles
-
Efficient ligation of the Schistosoma hammerhead ribozyme.Biochemistry. 2007 Mar 27;46(12):3826-34. doi: 10.1021/bi062077r. Epub 2007 Feb 24. Biochemistry. 2007. PMID: 17319693 Free PMC article.
-
Leakage and slow allostery limit performance of single drug-sensing aptazyme molecules based on the hammerhead ribozyme.RNA. 2009 Jan;15(1):76-84. doi: 10.1261/rna.1346609. Epub 2008 Nov 24. RNA. 2009. PMID: 19029309 Free PMC article.
-
Structural Simplicity and Mechanistic Complexity in the Hammerhead Ribozyme.Prog Mol Biol Transl Sci. 2018;159:177-202. doi: 10.1016/bs.pmbts.2018.07.006. Epub 2018 Sep 17. Prog Mol Biol Transl Sci. 2018. PMID: 30340787 Review.
-
Minimal Hammerhead Ribozymes with Uncompromised Catalytic Activity.J Mol Biol. 2015 Jul 17;427(14):2340-7. doi: 10.1016/j.jmb.2015.05.005. Epub 2015 May 14. J Mol Biol. 2015. PMID: 25981451 Free PMC article.
-
Ribozymes: from mechanistic studies to applications in vivo.J Biochem. 1995 Aug;118(2):251-8. doi: 10.1093/oxfordjournals.jbchem.a124899. J Biochem. 1995. PMID: 8543555 Review.
Cited by
-
Structural dynamics of a mitochondrial tRNA possessing weak thermodynamic stability.Biochemistry. 2014 Mar 11;53(9):1456-65. doi: 10.1021/bi401449z. Epub 2014 Feb 26. Biochemistry. 2014. PMID: 24520994 Free PMC article.
-
Folding of the hammerhead ribozyme: pyrrolo-cytosine fluorescence separates core folding from global folding and reveals a pH-dependent conformational change.RNA. 2012 Mar;18(3):434-48. doi: 10.1261/rna.030999.111. Epub 2012 Jan 24. RNA. 2012. PMID: 22274955 Free PMC article.
-
Single-molecule tools for enzymology, structural biology, systems biology and nanotechnology: an update.Arch Toxicol. 2014 Nov;88(11):1965-85. doi: 10.1007/s00204-014-1357-9. Epub 2014 Sep 12. Arch Toxicol. 2014. PMID: 25212907 Free PMC article. Review.
-
Enhanced hammerhead ribozyme turnover rates: Reevaluating therapeutic space for small catalytic RNAs.Mol Ther Nucleic Acids. 2024 Dec 21;36(1):102431. doi: 10.1016/j.omtn.2024.102431. eCollection 2025 Mar 11. Mol Ther Nucleic Acids. 2024. PMID: 40034206 Free PMC article.
-
Self-assembly Controls Self-cleavage of HHR from ASBVd (-): a Combined SANS and Modeling Study.Sci Rep. 2016 Jul 26;6:30287. doi: 10.1038/srep30287. Sci Rep. 2016. PMID: 27456224 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
