Structure, function, and folding of phosphoglycerate kinase are strongly perturbed by macromolecular crowding
- PMID: 20921368
- PMCID: PMC2955104
- DOI: 10.1073/pnas.1006760107
Structure, function, and folding of phosphoglycerate kinase are strongly perturbed by macromolecular crowding
Abstract
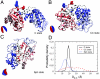
We combine experiment and computer simulation to show how macromolecular crowding dramatically affects the structure, function, and folding landscape of phosphoglycerate kinase (PGK). Fluorescence labeling shows that compact states of yeast PGK are populated as the amount of crowding agents (Ficoll 70) increases. Coarse-grained molecular simulations reveal three compact ensembles: C (crystal structure), CC (collapsed crystal), and Sph (spherical compact). With an adjustment for viscosity, crowded wild-type PGK and fluorescent PGK are about 15 times or more active in 200 mg/ml Ficoll than in aqueous solution. Our results suggest a previously undescribed solution to the classic problem of how the ADP and diphosphoglycerate binding sites of PGK come together to make ATP: Rather than undergoing a hinge motion, the ADP and substrate sites are already located in proximity under crowded conditions that mimic the in vivo conditions under which the enzyme actually operates. We also examine T-jump unfolding of PGK as a function of crowding experimentally. We uncover a nonmonotonic folding relaxation time vs. Ficoll concentration. Theory and modeling explain why an optimum concentration exists for fastest folding. Below the optimum, folding slows down because the unfolded state is stabilized relative to the transition state. Above the optimum, folding slows down because of increased viscosity.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
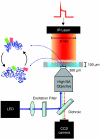
 .
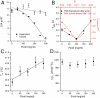
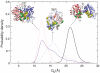
.  was computed using the size of the crowding agents equal to the native state of the protein. The simulations were scaled to match up the fastest folding rate observed in our experiments. (C) Melting temperature. (D) Cooperativity parameter Δg1 (free energy derivative) from Eq. 1. All error bars are ± 1 standard deviation.
was computed using the size of the crowding agents equal to the native state of the protein. The simulations were scaled to match up the fastest folding rate observed in our experiments. (C) Melting temperature. (D) Cooperativity parameter Δg1 (free energy derivative) from Eq. 1. All error bars are ± 1 standard deviation.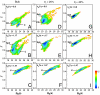
Comment in
-
Crowding and function reunite.Proc Natl Acad Sci U S A. 2010 Oct 12;107(41):17457-8. doi: 10.1073/pnas.1013095107. Epub 2010 Oct 6. Proc Natl Acad Sci U S A. 2010. PMID: 20926746 Free PMC article. No abstract available.
Similar articles
-
Protein stability and folding kinetics in the nucleus and endoplasmic reticulum of eucaryotic cells.Biophys J. 2011 Jul 20;101(2):421-30. doi: 10.1016/j.bpj.2011.05.071. Biophys J. 2011. PMID: 21767495 Free PMC article.
-
The extracellular protein VlsE is destabilized inside cells.J Mol Biol. 2014 Jan 9;426(1):11-20. doi: 10.1016/j.jmb.2013.08.024. Epub 2013 Sep 4. J Mol Biol. 2014. PMID: 24013077 Free PMC article.
-
An in vitro mimic of in-cell solvation for protein folding studies.Protein Sci. 2020 Apr;29(4):1060-1068. doi: 10.1002/pro.3833. Epub 2020 Feb 6. Protein Sci. 2020. PMID: 31994240 Free PMC article.
-
Denatured states of yeast phosphoglycerate kinase.Biochemistry (Mosc). 1998 Mar;63(3):259-75. Biochemistry (Mosc). 1998. PMID: 9526123 Review.
-
Phosphoglycerate kinase: structural aspects and functions, with special emphasis on the enzyme from Kinetoplastea.Open Biol. 2020 Nov;10(11):200302. doi: 10.1098/rsob.200302. Epub 2020 Nov 25. Open Biol. 2020. PMID: 33234025 Free PMC article. Review.
Cited by
-
Critical phenomena in the temperature-pressure-crowding phase diagram of a protein.Phys Rev X. 2019 Oct-Dec;9(4):041035. doi: 10.1103/physrevx.9.041035. Epub 2019 Nov 18. Phys Rev X. 2019. PMID: 32642303 Free PMC article.
-
Effects of External Perturbations on Protein Systems: A Microscopic View.ACS Omega. 2022 Nov 30;7(49):44556-44572. doi: 10.1021/acsomega.2c06199. eCollection 2022 Dec 13. ACS Omega. 2022. PMID: 36530249 Free PMC article. Review.
-
Effects of Macromolecular Crowding on the Conformational Ensembles of Disordered Proteins.J Phys Chem Lett. 2013 Oct 17;4(20):10.1021/jz401817x. doi: 10.1021/jz401817x. J Phys Chem Lett. 2013. PMID: 24312701 Free PMC article.
-
The parallel G-quadruplex structure of vertebrate telomeric repeat sequences is not the preferred folding topology under physiological conditions.Nucleic Acids Res. 2011 Jul;39(13):5768-75. doi: 10.1093/nar/gkr174. Epub 2011 Mar 30. Nucleic Acids Res. 2011. PMID: 21450807 Free PMC article.
-
Coupled protein diffusion and folding in the cell.PLoS One. 2014 Dec 1;9(12):e113040. doi: 10.1371/journal.pone.0113040. eCollection 2014. PLoS One. 2014. PMID: 25436502 Free PMC article.
References
-
- Gerstein M, Lesk AM, Chothia C. Structural mechanisms for domain movements in proteins. Biochemistry. 1994;33:6739–6749. - PubMed
-
- Blake C. Glycolysis—phosphotransfer hinges in PGK. Nature. 1997;385:204–205. - PubMed
-
- Bernstein BE, Michels PAM, Hol WGJ. Synergistic effects of substrate-induced conformational changes in phosphoglycerate kinase activation. Nature. 1997;385:275–278. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

