Initiation context modulates autoregulation of eukaryotic translation initiation factor 1 (eIF1)
- PMID: 20921384
- PMCID: PMC2964218
- DOI: 10.1073/pnas.1009269107
Initiation context modulates autoregulation of eukaryotic translation initiation factor 1 (eIF1)
Abstract
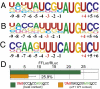
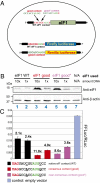
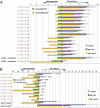
The central feature of standard eukaryotic translation initiation is small ribosome subunit loading at the 5' cap followed by its 5' to 3' scanning for a start codon. The preferred start is an AUG codon in an optimal context. Elaborate cellular machinery exists to ensure the fidelity of start codon selection. Eukaryotic initiation factor 1 (eIF1) plays a central role in this process. Here we show that the translation of eIF1 homologs in eukaryotes from diverse taxa involves initiation from an AUG codon in a poor context. Using human eIF1 as a model, we show that this poor context is necessary for an autoregulatory negative feedback loop in which a high level of eIF1 inhibits its own translation, establishing that variability in the stringency of start codon selection is used for gene regulation in eukaryotes. We show that the stringency of start codon selection (preferential utilization of optimal start sites) is increased to a surprising degree by overexpressing eIF1. The capacity for the cellular level of eIF1 to impact initiation through the variable stringency of initiation codon selection likely has significant consequences for the proteome in eukaryotes.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Stringency of start codon selection modulates autoregulation of translation initiation factor eIF5.Nucleic Acids Res. 2012 Apr;40(7):2898-906. doi: 10.1093/nar/gkr1192. Epub 2011 Dec 7. Nucleic Acids Res. 2012. PMID: 22156057 Free PMC article.
-
eIF1 discriminates against suboptimal initiation sites to prevent excessive uORF translation genome-wide.RNA. 2020 Apr;26(4):419-438. doi: 10.1261/rna.073536.119. Epub 2020 Jan 8. RNA. 2020. PMID: 31915290 Free PMC article.
-
eIF1 Loop 2 interactions with Met-tRNAi control the accuracy of start codon selection by the scanning preinitiation complex.Proc Natl Acad Sci U S A. 2018 May 1;115(18):E4159-E4168. doi: 10.1073/pnas.1800938115. Epub 2018 Apr 16. Proc Natl Acad Sci U S A. 2018. PMID: 29666249 Free PMC article.
-
Should I stay or should I go? Eukaryotic translation initiation factors 1 and 1A control start codon recognition.J Biol Chem. 2008 Oct 10;283(41):27345-27349. doi: 10.1074/jbc.R800031200. Epub 2008 Jun 30. J Biol Chem. 2008. PMID: 18593708 Free PMC article. Review.
-
The scanning mechanism of eukaryotic translation initiation.Annu Rev Biochem. 2014;83:779-812. doi: 10.1146/annurev-biochem-060713-035802. Epub 2014 Jan 29. Annu Rev Biochem. 2014. PMID: 24499181 Review.
Cited by
-
Prevalence of alternative AUG and non-AUG translation initiators and their regulatory effects across plants.Genome Res. 2020 Oct;30(10):1418-1433. doi: 10.1101/gr.261834.120. Epub 2020 Sep 24. Genome Res. 2020. PMID: 32973042 Free PMC article.
-
Non-AUG translation initiation in mammals.Genome Biol. 2022 May 9;23(1):111. doi: 10.1186/s13059-022-02674-2. Genome Biol. 2022. PMID: 35534899 Free PMC article. Review.
-
Discovery of paradoxical genes: reevaluating the prognostic impact of overexpressed genes in cancer.Front Cell Dev Biol. 2025 Jan 22;13:1525345. doi: 10.3389/fcell.2025.1525345. eCollection 2025. Front Cell Dev Biol. 2025. PMID: 39911323 Free PMC article. Review.
-
An essential fifth coding ORF in the sobemoviruses.Virology. 2013 Nov;446(1-2):397-408. doi: 10.1016/j.virol.2013.05.033. Epub 2013 Jul 2. Virology. 2013. PMID: 23830075 Free PMC article.
-
Functional analysis of the AUG initiator codon context reveals novel conserved sequences that disfavor mRNA translation in eukaryotes.Nucleic Acids Res. 2024 Feb 9;52(3):1064-1079. doi: 10.1093/nar/gkad1152. Nucleic Acids Res. 2024. PMID: 38038264 Free PMC article.
References
-
- Donahue TF. Genetic approaches to translation initiation. In: Sonenberg N, Hershey JWB, Mathews MB, editors. Saccharomyces cerevisiae. Translational Control of Gene Expression. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2000. pp. 595–614.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

