Dated molecular phylogenies indicate a Miocene origin for Arabidopsis thaliana
- PMID: 20921408
- PMCID: PMC2973009
- DOI: 10.1073/pnas.0909766107
Dated molecular phylogenies indicate a Miocene origin for Arabidopsis thaliana
Abstract
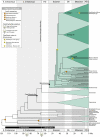
Dated molecular phylogenies are the basis for understanding species diversity and for linking changes in rates of diversification with historical events such as restructuring in developmental pathways, genome doubling, or dispersal onto a new continent. Valid fossil calibration points are essential to the accurate estimation of divergence dates, but for many groups of flowering plants fossil evidence is unavailable or limited. Arabidopsis thaliana, the primary genetic model in plant biology and the first plant to have its entire genome sequenced, belongs to one such group, the plant family Brassicaceae. Thus, the timing of A. thaliana evolution and the history of its genome have been controversial. We bring previously overlooked fossil evidence to bear on these questions and find the split between A. thaliana and Arabidopsis lyrata occurred about 13 Mya, and that the split between Arabidopsis and the Brassica complex (broccoli, cabbage, canola) occurred about 43 Mya. These estimates, which are two- to threefold older than previous estimates, indicate that gene, genomic, and developmental evolution occurred much more slowly than previously hypothesized and that Arabidopsis evolved during a period of warming rather than of cooling. We detected a 2- to 10-fold shift in species diversification rates on the branch uniting Brassicaceae with its sister families. The timing of this shift suggests a possible impact of the Cretaceous-Paleogene mass extinction on their radiation and that Brassicales codiversified with pierid butterflies that specialize on mustard-oil-producing plants.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Berr A, et al. Chromosome arrangement and nuclear architecture but not centromeric sequences are conserved between Arabidopsis thaliana and Arabidopsis lyrata. Plant J. 2006;48:771–783. - PubMed
-
- Koch MA, Haubold B, Mitchell-Olds T. Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis, and related genera (Brassicaceae) Mol Biol Evol. 2000;17:1483–1498. - PubMed
-
- Koch M, Haubold B, Mitchell-Olds T. Molecular systematics of the Brassicaceae: Evidence from coding plastidic matK and nuclear Chs sequences. Am J Bot. 2001;88:534–544. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

