One-step sample concentration, purification, and albumin depletion method for urinary proteomics
- PMID: 20923230
- PMCID: PMC2974758
- DOI: 10.1021/pr100924s
One-step sample concentration, purification, and albumin depletion method for urinary proteomics
Abstract
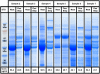
Workflows in urinary proteomics studies are often complex and require many steps to enrich, purify, deplete, and separate the complex mixture. Many of these methods are laborious, are time-consuming, and have the potential for error. Although individual steps of these methods have been previously studied, their downstream compatibilities with fractionation technologies such as off-gel electrophoresis have not been investigated. We developed a one-step sample preparation workflow that simultaneously (i) concentrates proteins, (ii) purifies by removing salts and other low molecular weight compounds, and (iii) depletes (albumin) from urine samples. This simple and robust workflow can be multiplexed and is compatible with a diverse range of downstream multidimensional separation technologies. Additionally, because of its high reproducibility and flexibility in processing samples with different volumes and concentrations, it has the potential to be used for standardization of urinary proteomics studies, as well as for studying other body fluids of similar complexity.
Figures





References
-
- O'Riordan E, Gross SS, Goligorsky MS. Technology Insight: renal proteomics--at the crossroads between promise and problems. Nat Clin Pract Nephrol. 2006;2(8):445–58. - PubMed
-
- Orenes-Pinero E, Corton M, Gonzalez-Peramato P, Algaba F, Casal I, Serrano A, Sanchez-Carbayo M. Searching urinary tumor markers for bladder cancer using a two-dimensional differential gel electrophoresis (2DDIGE) approach. J Proteome Res. 2007;6(11):4440–8. - PubMed
-
- Lafitte D, Dussol B, Andersen S, Vazi A, Dupuy P, Jensen ON, Berland Y, Verdier JM. Optimized preparation of urine samples for two-dimensional electrophoresis and initial application to patient samples. Clin Biochem. 2002;35(8):581–9. - PubMed
-
- Lee RS, Monigatti F, Lutchman M, Patterson T, Budnik B, Steen JA, Freeman MR, Steen H. Temporal variations of the postnatal rat urinary proteome as a reflection of systemic maturation. Proteomics. 2008;8(5):1097–112. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

