Clld7, a candidate tumor suppressor on chromosome 13q14, regulates pathways of DNA damage/repair and apoptosis
- PMID: 20926398
- PMCID: PMC2982930
- DOI: 10.1158/0008-5472.CAN-10-1960
Clld7, a candidate tumor suppressor on chromosome 13q14, regulates pathways of DNA damage/repair and apoptosis
Abstract
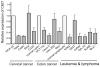
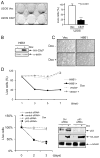
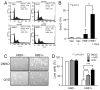
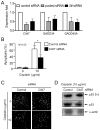
Chronic lymphocytic leukemia deletion gene 7 (Clld7) is a candidate tumor suppressor on chromosome 13q14. Clld7 encodes an evolutionarily conserved protein that contains an RCC1 domain plus broad complex, tramtrack, bric-a-brac (BTB), and POZ domains. In this study, we investigated the biological functions of Clld7 protein in inducible osteosarcoma cell lines. Clld7 induction inhibited cell growth, decreased cell viability, and increased γ-H2AX staining under conditions of caspase inhibition, indicating activation of the DNA damage/repair pathway. Real-time PCR analysis in tumor cells and normal human epithelial cells revealed Clld7 target genes that regulate DNA repair responses. Furthermore, depletion of Clld7 in normal human epithelial cells conferred resistance to apoptosis triggered by DNA damage. Taken together, the biological actions of Clld7 are consistent with those of a tumor suppressor.
Copyright © 2010 AACR.
Figures
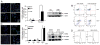

References
-
- Ripolles L, Ortega M, Ortuno F, et al. Genetic abnormalities and clinical outcome in chronic lymphocytic leukemia. Cancer Genet Cytogenet. 2006;171:57–64. - PubMed
-
- Cotter FE, Auer RL. Genetic alteration associated with chronic lymphocytic leukemia. Cytogenet Genome Res. 2007;118:310–9. - PubMed
-
- Wada M, Okamura T, Okada M, et al. Delineation of the frequently deleted region on chromosome arm 13q in B-cell non-Hodgkin’s lymphoma. Int J Hematol. 2000;71:159–66. - PubMed
-
- Wada M, Okamura T, Okada M, et al. Frequent chromosome arm 13q deletion in aggressive non-Hodgkin’s lymphoma. Leukemia. 1999;13:792–8. - PubMed
-
- Liu Y, Hermanson M, Grander D, et al. 13q deletions in lymphoid malignancies. Blood. 1995;86:1911–5. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

