Robust relationship inference in genome-wide association studies
- PMID: 20926424
- PMCID: PMC3025716
- DOI: 10.1093/bioinformatics/btq559
Robust relationship inference in genome-wide association studies
Abstract
Motivation: Genome-wide association studies (GWASs) have been widely used to map loci contributing to variation in complex traits and risk of diseases in humans. Accurate specification of familial relationships is crucial for family-based GWAS, as well as in population-based GWAS with unknown (or unrecognized) family structure. The family structure in a GWAS should be routinely investigated using the SNP data prior to the analysis of population structure or phenotype. Existing algorithms for relationship inference have a major weakness of estimating allele frequencies at each SNP from the entire sample, under a strong assumption of homogeneous population structure. This assumption is often untenable.
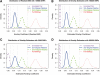
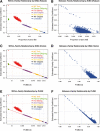
Results: Here, we present a rapid algorithm for relationship inference using high-throughput genotype data typical of GWAS that allows the presence of unknown population substructure. The relationship of any pair of individuals can be precisely inferred by robust estimation of their kinship coefficient, independent of sample composition or population structure (sample invariance). We present simulation experiments to demonstrate that the algorithm has sufficient power to provide reliable inference on millions of unrelated pairs and thousands of relative pairs (up to 3rd-degree relationships). Application of our robust algorithm to HapMap and GWAS datasets demonstrates that it performs properly even under extreme population stratification, while algorithms assuming a homogeneous population give systematically biased results. Our extremely efficient implementation performs relationship inference on millions of pairs of individuals in a matter of minutes, dozens of times faster than the most efficient existing algorithm known to us.
Availability: Our robust relationship inference algorithm is implemented in a freely available software package, KING, available for download at http://people.virginia.edu/∼wc9c/KING.
Figures


References
-
- Abecasis GR, et al. GRR: graphical representation of relationship errors. Bioinformatics. 2001;17:742–743. - PubMed
-
- Abecasis GR, et al. Merlin–rapid analysis of dense genetic maps using sparse gene flow trees. Nat. Genet. 2002;30:97–101. - PubMed
-
- Chen WM, Deng HW. A general and accurate approach for computing the statistical power of the transmission disequilibrium test for complex disease genes. Genet. Epidemiol. 2001;21:53–67. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

