An unprecedented nucleic acid capture mechanism for excision of DNA damage
- PMID: 20927102
- PMCID: PMC4160814
- DOI: 10.1038/nature09428
An unprecedented nucleic acid capture mechanism for excision of DNA damage
Abstract
DNA glycosylases that remove alkylated and deaminated purine nucleobases are essential DNA repair enzymes that protect the genome, and at the same time confound cancer alkylation therapy, by excising cytotoxic N3-methyladenine bases formed by DNA-targeting anticancer compounds. The basis for glycosylase specificity towards N3- and N7-alkylpurines is believed to result from intrinsic instability of the modified bases and not from direct enzyme functional group chemistry. Here we present crystal structures of the recently discovered Bacillus cereus AlkD glycosylase in complex with DNAs containing alkylated, mismatched and abasic nucleotides. Unlike other glycosylases, AlkD captures the extrahelical lesion in a solvent-exposed orientation, providing an illustration for how hydrolysis of N3- and N7-alkylated bases may be facilitated by increased lifetime out of the DNA helix. The structures and supporting biochemical analysis of base flipping and catalysis reveal how the HEAT repeats of AlkD distort the DNA backbone to detect non-Watson-Crick base pairs without duplex intercalation.
Figures

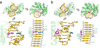
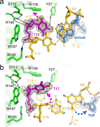
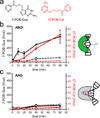

References
-
- Friedberg EC, et al. DNA repair: from molecular mechanism to human disease. DNA repair. 2006;5(8):986–996. - PubMed
-
- Singer B, Grunberger D. Molecular Biology of Mutagens and Carcinogens: Intrinsic Properties of Nucleic Acids. Plenum Press: New York; 1983.
-
- Holt S, Yen TY, Sangaiah R, Swenberg JA. Detection of 1,N6-ethenoadenine in rat urine after chloroethylene oxide exposure. Carcinogenesis. 1998;19(10):1763–1769. - PubMed
-
- Shuker DE, Bailey E, Parry A, Lamb J, Farmer PB. The determination of urinary 3-methyladenine in humans as a potential monitor of exposure to methylating agents. Carcinogenesis. 1987;8(7):959–962. - PubMed
-
- Shuker DE, Farmer PB. Relevance of urinary DNA adducts as markers of carcinogen exposure. Chem Res Toxicol. 1992;5(4):450–460. - PubMed
References to Methods
-
- Irani RJ, SantaLucia J. Jr. The synthesis of anti-fixed 3-methyl-3-deaza-2'- deoxyadenosine and other 3H-imidazo[4,5-c]pyridine analogs. Nucleosides Nucleotides Nucleic Acids. 2002;21:737–751. - PubMed
-
- Otwinowski Z, Minor W. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. - PubMed
-
- McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr D Biol Crystallogr. 2005;61:458–464. - PubMed
-
- Brunger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54(Pt 5):905–921. - PubMed
-
- McRee DE. XtalView/Xfit--A versatile program for manipulating atomic coordinates and electron density. Journal of structural biology. 1999;125:156–165. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

