Mitochondrial genome evolution in fire ants (Hymenoptera: Formicidae)
- PMID: 20929580
- PMCID: PMC2958920
- DOI: 10.1186/1471-2148-10-300
Mitochondrial genome evolution in fire ants (Hymenoptera: Formicidae)
Abstract
Background: Complete mitochondrial genome sequences have become important tools for the study of genome architecture, phylogeny, and molecular evolution. Despite the rapid increase in available mitogenomes, the taxonomic sampling often poorly reflects phylogenetic diversity and is often also biased to represent deeper (family-level) evolutionary relationships.
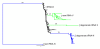
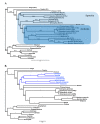
Results: We present the first fully sequenced ant (Hymenoptera: Formicidae) mitochondrial genomes. We sampled four mitogenomes from three species of fire ants, genus Solenopsis, which represent various evolutionary depths. Overall, ant mitogenomes appear to be typical of hymenopteran mitogenomes, displaying a general A+T-bias. The Solenopsis mitogenomes are slightly more compact than other hymentoperan mitogenomes (~15.5 kb), retaining all protein coding genes, ribosomal, and transfer RNAs. We also present evidence of recombination between the mitogenomes of the two conspecific Solenopsis mitogenomes. Finally, we discuss potential ways to improve the estimation of phylogenies using complete mitochondrial genome sequences.
Conclusions: The ant mitogenome presents an important addition to the continued efforts in studying hymenopteran mitogenome architecture, evolution, and phylogenetics. We provide further evidence that the sampling across many taxonomic levels (including conspecifics and congeners) is useful and important to gain detailed insights into mitogenome evolution. We also discuss ways that may help improve the use of mitogenomes in phylogenetic analyses by accounting for non-stationary and non-homogeneous evolution among branches.
Figures




References
-
- Cameron SL, Lambkin CL, Barker SC, Whiting MF. A mitochondrial genome phylogeny of Diptera: whole genome sequence data accurately resolve relationships over broad timescales with high precision. Syst Entomol. 2007;32:40–59. doi: 10.1111/j.1365-3113.2006.00355.x. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

