A general mechanism for network-dosage compensation in gene circuits
- PMID: 20929850
- PMCID: PMC3138731
- DOI: 10.1126/science.1190544
A general mechanism for network-dosage compensation in gene circuits
Abstract
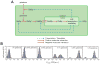
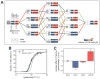
Coping with variations in network dosage is crucial for maintaining optimal function in gene networks. We explored how network structure facilitates network-level dosage compensation. By using the yeast galactose network as a model, we combinatorially deleted one of the two copies of its four regulatory genes and found that network activity was robust to the change in network dosage. A mathematical analysis revealed that a two-component genetic circuit with elements of opposite regulatory activity (activator and inhibitor) constitutes a minimal requirement for network-dosage invariance. Specific interaction topologies and a one-to-one interaction stoichiometry between the activating and inhibiting agents were additional essential elements facilitating dosage invariance. This mechanism of network-dosage invariance could represent a general design for gene network structure in cells.
Figures
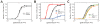

References
-
- Lee JA, Lupski JR. Genomic rearrangements and gene copy-number alterations as a cause of nervous system disorders. Neuron. 2006;52:103. - PubMed
-
- Galitski T, Saldanha AJ, Styles CA, Lander ES, Fink GR. Ploidy regulation of gene expression. Science. 1999;285:251. - PubMed
-
- Kellis M, Birren BW, Lander ES. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature. 2004;428:617. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

