Uncovering the human methyltransferasome
- PMID: 20930037
- PMCID: PMC3013446
- DOI: 10.1074/mcp.M110.000976
Uncovering the human methyltransferasome
Abstract
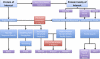
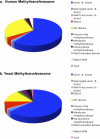
We present a comprehensive analysis of the human methyltransferasome. Primary sequences, predicted secondary structures, and solved crystal structures of known methyltransferases were analyzed by hidden Markov models, Fisher-based statistical matrices, and fold recognition prediction-based threading algorithms to create a model, or profile, of each methyltransferase superfamily. These profiles were used to scan the human proteome database and detect novel methyltransferases. 208 proteins in the human genome are now identified as known or putative methyltransferases, including 38 proteins that were not annotated previously. To date, 30% of these proteins have been linked to disease states. Possible substrates of methylation for all of the SET domain and SPOUT methyltransferases as well as 100 of the 131 seven-β-strand methyltransferases were surmised from sequence similarity clusters based on alignments of the substrate-specific domains.
Figures



References
-
- Cheng X., Blumenthal R. M. (eds) (1999) S-Adenosylmethionine-dependent Methyltransferases: Structures and Functions, World Scientific, Singapore
-
- Martin J. L., McMillan F. M. (2002) SAM (dependent) I AM: the S-adenosylmethionine-dependent methyltransferase fold. Curr. Opin. Struct. Biol. 12, 783–793 - PubMed
-
- Katz J. E., Dlakić M., Clarke S. (2003) Automated identification of putative methyltransferases from genomic open reading frames. Mol. Cell. Proteomics 2, 525–540 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

