A transcriptional map of the impact of endurance exercise training on skeletal muscle phenotype
- PMID: 20930125
- PMCID: PMC3253010
- DOI: 10.1152/japplphysiol.00634.2010
A transcriptional map of the impact of endurance exercise training on skeletal muscle phenotype
Abstract
The molecular pathways that are activated and contribute to physiological remodeling of skeletal muscle in response to endurance exercise have not been fully characterized. We previously reported that ∼800 gene transcripts are regulated following 6 wk of supervised endurance training in young sedentary males, referred to as the training-responsive transcriptome (TRT) (Timmons JA et al. J Appl Physiol 108: 1487-1496, 2010). Here we utilized this database together with data on biological variation in muscle adaptation to aerobic endurance training in both humans and a novel out-bred rodent model to study the potential regulatory molecules that coordinate this complex network of genes. We identified three DNA sequences representing RUNX1, SOX9, and PAX3 transcription factor binding sites as overrepresented in the TRT. In turn, miRNA profiling indicated that several miRNAs targeting RUNX1, SOX9, and PAX3 were downregulated by endurance training. The TRT was then examined by contrasting subjects who demonstrated the least vs. the greatest improvement in aerobic capacity (low vs. high responders), and at least 100 of the 800 TRT genes were differentially regulated, thus suggesting regulation of these genes may be important for improving aerobic capacity. In high responders, proangiogenic and tissue developmental networks emerged as key candidates for coordinating tissue aerobic adaptation. Beyond RNA-level validation there were several DNA variants that associated with maximal aerobic capacity (Vo(₂max)) trainability in the HERITAGE Family Study but these did not pass conservative Bonferroni adjustment. In addition, in a rat model selected across 10 generations for high aerobic training responsiveness, we found that both the TRT and a homologous subset of the human high responder genes were regulated to a greater degree in high responder rodent skeletal muscle. This analysis provides a comprehensive map of the transcriptomic features important for aerobic exercise-induced improvements in maximal oxygen consumption.
Figures
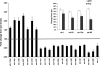
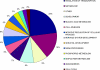


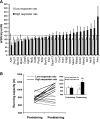
Comment in
-
Running with regulation.J Appl Physiol (1985). 2011 Jan;110(1):13-4. doi: 10.1152/japplphysiol.01327.2010. Epub 2010 Nov 11. J Appl Physiol (1985). 2011. PMID: 21071585 No abstract available.
References
-
- Alexa A, Rahnenfuhrer J, Lengauer T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 22: 1600–1607, 2006 - PubMed
-
- An P, Teran-Garcia M, Rice T, Rankinen T, Weisnagel SJ, Bergman RN, Boston RC, Mandel S, Stefanovski D, Leon AS, Skinner JS, Rao DC, Bouchard C. Genome-wide linkage scans for prediabetes phenotypes in response to 20 weeks of endurance exercise training in non-diabetic whites and blacks: the HERITAGE Family Study. Diabetologia 48: 1142–1149, 2005 - PubMed
-
- Baranowsky A, Mokkapati S, Bechtel M, Krugel J, Miosge N, Wickenhauser C, Smyth N, Nischt R. Impaired wound healing in mice lacking the basement membrane protein nidogen 1. Matrix Biol 29: 15–21, 2010 - PubMed
-
- Bonauer A, Carmona G, Iwasaki M, Mione M, Koyanagi M, Fischer A, Burchfield J, Fox H, Doebele C, Ohtani K, Chavakis E, Potente M, Tjwa M, Urbich C, Zeiher AM, Dimmeler S. MicroRNA-92a controls angiogenesis and functional recovery of ischemic tissues in mice. Science 324: 1710–1713, 2009 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

