The C-terminus of nucleolin promotes the formation of the c-MYC G-quadruplex and inhibits c-MYC promoter activity
- PMID: 20932061
- PMCID: PMC2976822
- DOI: 10.1021/bi100509s
The C-terminus of nucleolin promotes the formation of the c-MYC G-quadruplex and inhibits c-MYC promoter activity
Abstract
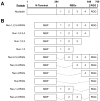
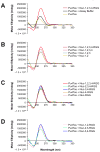
Nucleolin, the most abundant nucleolar phosphoprotein of eukaryotic cells, is known primarily for its role in ribosome biogenesis and cell proliferation. It is, however, a multifunctional protein that, depending on the cellular context, can drive either cell proliferation or apoptosis. Our laboratory recently demonstrated that nucleolin can function as a repressor of c-MYC transcription by binding to and stabilizing the formation of a G-quadruplex structure in a region of the c-MYC promoter responsible for controlling 85-90% of c-MYC's transcriptional activity. In this study, we investigate the structural elements of nucleolin that are required for c-MYC repression. The effect of nucleolin deletion mutants on the formation and stability of the c-MYC G-quadruplex, as well as c-MYC transcriptional activity, was assessed by circular dichroism spectropolarimetry, thermal stability, and in vitro transcription. Here we report that nucleolin's RNA binding domains 3 and 4, as well as the arginine-glycine-glycine (RGG) domain, are required to repress c-MYC transcription.
Figures




Similar articles
-
Identification and characterization of nucleolin as a c-myc G-quadruplex-binding protein.J Biol Chem. 2009 Aug 28;284(35):23622-35. doi: 10.1074/jbc.M109.018028. Epub 2009 Jul 6. J Biol Chem. 2009. PMID: 19581307 Free PMC article.
-
Truncated G-Quadruplex Isomers Cross-Talk with the Transcription Factors To Maintain Homeostatic Equilibria in c-MYC Transcription.Biochemistry. 2019 Apr 16;58(15):1975-1991. doi: 10.1021/acs.biochem.9b00030. Epub 2019 Apr 5. Biochemistry. 2019. PMID: 30920805
-
Structural basis for nucleolin recognition of MYC promoter G-quadruplex.Science. 2025 Apr 18;388(6744):eadr1752. doi: 10.1126/science.adr1752. Epub 2025 Apr 18. Science. 2025. PMID: 40245140
-
Molecular dissection of nucleolin's role in growth and cell proliferation: new insights.FASEB J. 1999 Nov;13(14):1911-22. FASEB J. 1999. PMID: 10544174 Review.
-
Structure and functions of nucleolin.J Cell Sci. 1999 Mar;112 ( Pt 6):761-72. doi: 10.1242/jcs.112.6.761. J Cell Sci. 1999. PMID: 10036227 Review.
Cited by
-
G-quadruplex ligands targeting telomeres do not inhibit HIV promoter activity and cooperate with latency reversing agents in killing latently infected cells.Cell Cycle. 2020 Sep;19(18):2298-2313. doi: 10.1080/15384101.2020.1796268. Epub 2020 Aug 17. Cell Cycle. 2020. PMID: 32807015 Free PMC article.
-
Binding of cellular nucleolin with the viral core RNA G-quadruplex structure suppresses HCV replication.Nucleic Acids Res. 2019 Jan 10;47(1):56-68. doi: 10.1093/nar/gky1177. Nucleic Acids Res. 2019. PMID: 30462330 Free PMC article.
-
Nucleolin protein interacts with microprocessor complex to affect biogenesis of microRNAs 15a and 16.J Biol Chem. 2011 Dec 23;286(51):44095-44103. doi: 10.1074/jbc.M111.265439. Epub 2011 Nov 2. J Biol Chem. 2011. PMID: 22049078 Free PMC article.
-
Higher-order G-quadruplexes in promoters are untapped drug targets.Front Chem. 2023 Jun 7;11:1211512. doi: 10.3389/fchem.2023.1211512. eCollection 2023. Front Chem. 2023. PMID: 37351517 Free PMC article. Review.
-
Parallel G-quadruplexes recruit the HSV-1 transcription factor ICP4 to promote viral transcription in herpes virus-infected human cells.Commun Biol. 2021 Apr 30;4(1):510. doi: 10.1038/s42003-021-02035-y. Commun Biol. 2021. PMID: 33931711 Free PMC article.
References
-
- Wierstra I, Alves J. The c-myc promoter: still MysterY and Challenge. Adv Cancer Res. 2008;99:113–333. - PubMed
-
- Davis JT. G-quartets 40 years later: from 5′-GMP to molecular biology and supramolecular chemistry. Angew Chem Int Ed Engl. 2004;43:668–698. - PubMed
-
- Ghosal G, Muniyappa K. Hoogsteen base-pairing revisited: resolving a role in normal biological processes and human diseases. Biochem Biophys Res Commun. 2006;343:1–7. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

