Analysis of X-ray structures of matrix metalloproteinases via chaotic map clustering
- PMID: 20932281
- PMCID: PMC3098083
- DOI: 10.1186/1471-2105-11-500
Analysis of X-ray structures of matrix metalloproteinases via chaotic map clustering
Abstract
Background: Matrix metalloproteinases (MMPs) are well-known biological targets implicated in tumour progression, homeostatic regulation, innate immunity, impaired delivery of pro-apoptotic ligands, and the release and cleavage of cell-surface receptors. With this in mind, the perception of the intimate relationships among diverse MMPs could be a solid basis for accelerated learning in designing new selective MMP inhibitors. In this regard, decrypting the latent molecular reasons in order to elucidate similarity among MMPs is a key challenge.
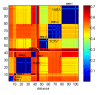
Results: We describe a pairwise variant of the non-parametric chaotic map clustering (CMC) algorithm and its application to 104 X-ray MMP structures. In this analysis electrostatic potentials are computed and used as input for the CMC algorithm. It was shown that differences between proteins reflect genuine variation of their electrostatic potentials. In addition, the analysis has been also extended to analyze the protein primary structures and the molecular shapes of the MMP co-crystallised ligands.
Conclusions: The CMC algorithm was shown to be a valuable tool in knowledge acquisition and transfer from MMP structures. Based on the variation of electrostatic potentials, CMC was successful in analysing the MMP target family landscape and different subsites. The first investigation resulted in rational figure interpretation of both domain organization as well as of substrate specificity classifications. The second made it possible to distinguish the MMP classes, demonstrating the high specificity of the S1' pocket, to detect both the occurrence of punctual mutations of ionisable residues and different side-chain conformations that likely account for induced-fit phenomena. In addition, CMC demonstrated a potential comparable to the most popular UPGMA (Unweighted Pair Group Method with Arithmetic mean) method that, at present, represents a standard clustering bioinformatics approach. Interestingly, CMC and UPGMA resulted in closely comparable outcomes, but often CMC produced more informative and more easy interpretable dendrograms. Finally, CMC was successful for standard pairwise analysis (i.e., Smith-Waterman algorithm) of protein sequences and was used to convincingly explain the complementarity existing between the molecular shapes of the co-crystallised ligand molecules and the accessible MMP void volumes.
Figures





Similar articles
-
The intermediate S1' pocket of the endometase/matrilysin-2 active site revealed by enzyme inhibition kinetic studies, protein sequence analyses, and homology modeling.J Biol Chem. 2003 Dec 19;278(51):51646-53. doi: 10.1074/jbc.M310109200. Epub 2003 Oct 7. J Biol Chem. 2003. PMID: 14532275
-
Specificity of binding with matrix metalloproteinases.Exp Suppl. 2012;103:35-56. doi: 10.1007/978-3-0348-0364-9_2. Exp Suppl. 2012. PMID: 22642189 Review.
-
Screening of matrix metalloproteinases available from the protein data bank: insights into biological functions, domain organization, and zinc binding groups.J Chem Inf Model. 2007 Nov-Dec;47(6):2439-48. doi: 10.1021/ci700119r. Epub 2007 Oct 24. J Chem Inf Model. 2007. PMID: 17958346
-
Active site specificity profiling of the matrix metalloproteinase family: Proteomic identification of 4300 cleavage sites by nine MMPs explored with structural and synthetic peptide cleavage analyses.Matrix Biol. 2016 Jan;49:37-60. doi: 10.1016/j.matbio.2015.09.003. Epub 2015 Sep 25. Matrix Biol. 2016. PMID: 26407638
-
Structural basis of the matrix metalloproteinases and their physiological inhibitors, the tissue inhibitors of metalloproteinases.Biol Chem. 2003 Jun;384(6):863-72. doi: 10.1515/BC.2003.097. Biol Chem. 2003. PMID: 12887053 Review.
Cited by
-
Insights into the complex formed by matrix metalloproteinase-2 and alloxan inhibitors: molecular dynamics simulations and free energy calculations.PLoS One. 2011;6(10):e25597. doi: 10.1371/journal.pone.0025597. Epub 2011 Oct 5. PLoS One. 2011. PMID: 21998672 Free PMC article.
-
Mammographic images segmentation based on chaotic map clustering algorithm.BMC Med Imaging. 2014 Mar 25;14:12. doi: 10.1186/1471-2342-14-12. BMC Med Imaging. 2014. PMID: 24666766 Free PMC article.
References
-
- Burzlaff N. Concepts and Models in Bioinorganic Chemistry. Wiley-Vch; 2006. From Model Complexes for Zinc-Containing Enzymes; pp. 397–429.
-
- Willett P. Similarity and Clustering in Chemical Information Systems. New York: John Wiley & Sons; 1987.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

