Improved variant discovery through local re-alignment of short-read next-generation sequencing data using SRMA
- PMID: 20932289
- PMCID: PMC3218665
- DOI: 10.1186/gb-2010-11-10-r99
Improved variant discovery through local re-alignment of short-read next-generation sequencing data using SRMA
Abstract
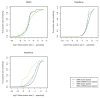
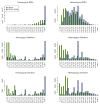
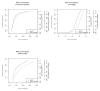
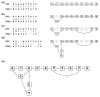
A primary component of next-generation sequencing analysis is to align short reads to a reference genome, with each read aligned independently. However, reads that observe the same non-reference DNA sequence are highly correlated and can be used to better model the true variation in the target genome. A novel short-read micro realigner, SRMA, that leverages this correlation to better resolve a consensus of the underlying DNA sequence of the targeted genome is described here.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

