Enzymes of the mevalonate pathway of isoprenoid biosynthesis
- PMID: 20932952
- PMCID: PMC3026612
- DOI: 10.1016/j.abb.2010.09.028
Enzymes of the mevalonate pathway of isoprenoid biosynthesis
Abstract
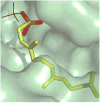
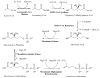
The mevalonate pathway accounts for conversion of acetyl-CoA to isopentenyl 5-diphosphate, the versatile precursor of polyisoprenoid metabolites and natural products. The pathway functions in most eukaryotes, archaea, and some eubacteria. Only recently has much of the functional and structural basis for this metabolism been reported. The biosynthetic acetoacetyl-CoA thiolase and HMG-CoA synthase reactions rely on key amino acids that are different but are situated in active sites that are similar throughout the family of initial condensation enzymes. Both bacterial and animal HMG-CoA reductases have been extensively studied and the contrasts between these proteins and their interactions with statin inhibitors defined. The conversion of mevalonic acid to isopentenyl 5-diphosphate involves three ATP-dependent phosphorylation reactions. While bacterial enzymes responsible for these three reactions share a common protein fold, animal enzymes differ in this respect as the recently reported structure of human phosphomevalonate kinase demonstrates. There are significant contrasts between observations on metabolite inhibition of mevalonate phosphorylation in bacteria and animals. The structural basis for these contrasts has also recently been reported. Alternatives to the phosphomevalonate kinase and mevalonate diphosphate decarboxylase reactions may exist in archaea. Thus, new details regarding isopentenyl diphosphate synthesis from acetyl-CoA continue to emerge.
Copyright © 2010 Elsevier Inc. All rights reserved.
Figures



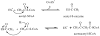

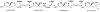
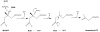
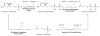
References
-
- Bloch K. Science. 1965;150:19–28. - PubMed
-
- Bochar FJDA, Stauffacher CV, Rodwell VW. Biosynthesis of mevalonic acid from acetyl-CoA. Pergamon Press; New York: 1999.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

