An RNA conformational switch regulates pre-18S rRNA cleavage
- PMID: 20934433
- PMCID: PMC3006654
- DOI: 10.1016/j.jmb.2010.09.064
An RNA conformational switch regulates pre-18S rRNA cleavage
Abstract
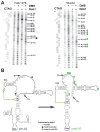
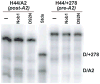
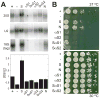
To produce mature ribosomal RNAs (rRNAs), polycistronic rRNA transcripts are cleaved in an ordered series of events. We have uncovered the molecular basis for the ordering of two essential cleavage steps at the 3'-end of 18S rRNA. Using in vitro and in vivo structure probing, RNA binding and cleavage experiments, and yeast genetics, we demonstrate that a conserved RNA sequence in the spacer region between the 18S and 5.8S rRNAs base-pairs with the decoding site of 18S rRNA in early assembly intermediates. Nucleolar cleavage at site A(2) excises this sequence element, leading to a conformational switch in pre-18S rRNA, by which the ribosomal decoding site is formed. This conformational switch positions the nuclease Nob1 for cytoplasmic cleavage at the 3'-end of 18S rRNA and is required for the final maturation step of 18S rRNA in vivo and in vitro. More generally, our data show that the intrinsic ability of RNA to form stable structural switches is exploited to order and regulate RNA-dependent biological processes.
Copyright © 2010 Elsevier Ltd. All rights reserved.
Figures


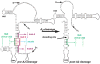
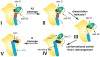
Comment in
-
18S rRNA: a tale of the tail.J Mol Biol. 2011 Jan 7;405(1):1-2. doi: 10.1016/j.jmb.2010.10.022. Epub 2010 Oct 20. J Mol Biol. 2011. PMID: 20969874 Free PMC article. No abstract available.
Similar articles
-
Base pairing between U3 small nucleolar RNA and the 5' end of 18S rRNA is required for pre-rRNA processing.Mol Cell Biol. 1999 Sep;19(9):6012-9. doi: 10.1128/MCB.19.9.6012. Mol Cell Biol. 1999. PMID: 10454548 Free PMC article.
-
Imp3 unfolds stem structures in pre-rRNA and U3 snoRNA to form a duplex essential for small subunit processing.RNA. 2013 Oct;19(10):1372-83. doi: 10.1261/rna.039511.113. Epub 2013 Aug 26. RNA. 2013. PMID: 23980203 Free PMC article.
-
Ribosomal RNA processing in Candida albicans.RNA. 2011 Dec;17(12):2235-48. doi: 10.1261/rna.028050.111. Epub 2011 Oct 25. RNA. 2011. PMID: 22028364 Free PMC article.
-
Two distinct recognition signals define the site of endonucleolytic cleavage at the 5'-end of yeast 18S rRNA.EMBO J. 1995 Oct 2;14(19):4883-92. doi: 10.1002/j.1460-2075.1995.tb00169.x. EMBO J. 1995. PMID: 7588617 Free PMC article.
-
Nob1 binds the single-stranded cleavage site D at the 3'-end of 18S rRNA with its PIN domain.Proc Natl Acad Sci U S A. 2009 Aug 25;106(34):14259-64. doi: 10.1073/pnas.0905403106. Epub 2009 Aug 14. Proc Natl Acad Sci U S A. 2009. PMID: 19706509 Free PMC article.
Cited by
-
Ribosomal RNA Biogenesis and Its Response to Chilling Stress in Oryza sativa.Plant Physiol. 2018 May;177(1):381-397. doi: 10.1104/pp.17.01714. Epub 2018 Mar 19. Plant Physiol. 2018. PMID: 29555785 Free PMC article.
-
Cofactor-dependent specificity of a DEAD-box protein.Proc Natl Acad Sci U S A. 2013 Jul 16;110(29):E2668-76. doi: 10.1073/pnas.1302577110. Epub 2013 Apr 29. Proc Natl Acad Sci U S A. 2013. PMID: 23630256 Free PMC article.
-
Hierarchical recruitment into nascent ribosomes of assembly factors required for 27SB pre-rRNA processing in Saccharomyces cerevisiae.Nucleic Acids Res. 2012 Sep 1;40(17):8646-61. doi: 10.1093/nar/gks609. Epub 2012 Jun 26. Nucleic Acids Res. 2012. PMID: 22735702 Free PMC article.
-
Structural and functional analysis of the archaeal endonuclease Nob1.Nucleic Acids Res. 2012 Apr;40(7):3259-74. doi: 10.1093/nar/gkr1186. Epub 2011 Dec 10. Nucleic Acids Res. 2012. PMID: 22156373 Free PMC article.
-
A translation-like cycle is a quality control checkpoint for maturing 40S ribosome subunits.Cell. 2012 Jul 6;150(1):111-21. doi: 10.1016/j.cell.2012.04.044. Cell. 2012. PMID: 22770215 Free PMC article.
References
-
- Karbstein K, Jonas S, Doudna JA. An essential GTPase promotes assembly of preribosomal RNA processing complexes. Mol Cell. 2005;20:633–43. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

