Mutator phenotypes due to DNA replication infidelity
- PMID: 20934516
- PMCID: PMC3087159
- DOI: 10.1016/j.semcancer.2010.10.003
Mutator phenotypes due to DNA replication infidelity
Abstract
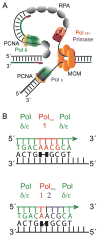
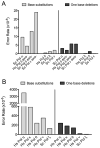
This article considers the fidelity of DNA replication performed by eukaryotic DNA polymerases involved in replicating the nuclear genome. DNA replication fidelity can vary widely depending on the DNA polymerase, the composition of the error, the flanking sequence, the presence of DNA damage and the ability to correct errors. As a consequence, defects in processes that determine DNA replication fidelity can confer strong mutator phenotypes whose specificity can help determine the molecular nature of the defect.
Published by Elsevier Ltd.
Conflict of interest statement
The authors declare that there is no conflict of interest.
Figures

References
-
- Bebenek K, Kunkel TA. Functions of DNA polymerases. Adv Protein Chem. 2004;69:137–65. - PubMed
-
- Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA repair and mutagenesis. 2. ASM Press; 2006.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

