Co-activation of epidermal growth factor receptor and c-MET defines a distinct subset of lung adenocarcinomas
- PMID: 20934974
- PMCID: PMC2966779
- DOI: 10.2353/ajpath.2010.100217
Co-activation of epidermal growth factor receptor and c-MET defines a distinct subset of lung adenocarcinomas
Abstract
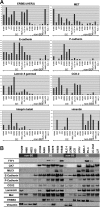
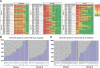
Epidermal growth factor receptor (EGFR) and MET are molecular targets for lung cancer treatment. The relationships between expression, activation, and gene abnormalities of these two targets are currently unclear. Here, we demonstrate that a panel of 40 lung cancer cell lines could be classified into two groups. Group I was characterized by (1) high phosphorylations of MET and EGFR, (2) frequent mutation or amplification of EGFR, MET, and human epidermal growth factor receptor-2 (HER2), (3) high expressions of bronchial epithelial markers (thyroid transcription factor-1 (TTF-1), MUC1, and Cytokeratin 7 (CK7)); and (4) high expressions of MET, human epidermal growth factor receptor-3, E-cadherin, cyclooxygenase-2, and laminin gamma2. In contrast, Group II exhibited little or no phosphorylation of MET and EGFR; no mutation or amplification of EGFR, MET, and HER2; were triple-negative for TTF-1, MUC1, and CK7; and showed high expressions of vimentin, fibroblast growth factor receptor-1, and transcription factor 8. Importantly, Group I was more sensitive to gefitinib and more resistant to cisplatin and paclitaxel than Group II. The clinical relevance was confirmed in publicly available data on 442 primary lung adenocarcinoma patients; survival benefits by postoperative chemotherapy were seen in only patients with tumors corresponding to Group II. Overall, co-activation of EGFR and MET defines a distinct subgroup of lung carcinoma with characteristic genetic abnormalities, gene expression pattern, and response to chemotherapeutic reagents.
Figures



References
-
- Tokyo: Ministry of Health, Labor, and Welfare; Statistics and Information Department. Vital Statistics, 2000. 2001
-
- Jemal A, Siegel R, Ward E, Murray T, Xu J, Smigal C, Thun MJ. Cancer Statistics, 2006. CA Cancer J Clin. 2006;56:106–130. - PubMed
-
- Naruke T, Tsuchiya R, Kondo H, Asamura H. Prognosis and survival after resection for bronchogenic carcinoma based on the 1997 TNM-staging classification: the Japanese experience. Ann Thorac Surg. 2001;71:1759–1764. - PubMed
-
- Janssen-Heijnen ML, Coebergh JW. Trends in incidence and prognosis of the histological subtypes of lung cancer in North America. Australia, New Zealand and Europe. Lung Cancer. 2001;31:123–137. - PubMed
-
- Riely GJ, Politi KA, Miller VA, Pao W. Update on epidermal growth factor receptor mutations in non-small cell lung cancer. Clin Cancer Res. 2006;12:7232–7241. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

