End-processing during non-homologous end-joining: a role for exonuclease 1
- PMID: 20935051
- PMCID: PMC3035470
- DOI: 10.1093/nar/gkq886
End-processing during non-homologous end-joining: a role for exonuclease 1
Abstract
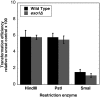
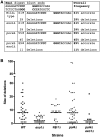
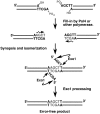
Non-homologous end-joining (NHEJ) is a critical error-prone pathway of double strand break repair. We recently showed that tyrosyl DNA phosphodiesterase 1 (Tdp1) regulates the accuracy of NHEJ repair junction formation in yeast. We assessed the role of other enzymes in the accuracy of junction formation using a plasmid repair assay. We found that exonuclease 1 (Exo1) is important in assuring accurate junction formation during NHEJ. Like tdp1Δ mutants, exo1Δ yeast cells repairing plasmids with 5'-extensions can produce repair junctions with templated insertions. We also found that exo1Δ mutants have a reduced median size of deletions when joining DNA with blunt ends. Surprisingly, exo1Δ pol4Δ mutants repair blunt ends with a very low frequency of deletions. This result suggests that there are multiple pathways that process blunt ends prior to end-joining. We propose that Exo1 acts at a late stage in end-processing during NHEJ. Exo1 can reverse nucleotide additions occurring due to polymerization, and may also be important for processing ends to expose microhomologies needed for NHEJ. We propose that accurate joining is controlled at two steps, a first step that blocks modification of DNA ends, which requires Tdp1, and a second step that occurs after synapsis that requires Exo1.
Figures




References
-
- Taccioli GE, Gottlieb TM, Blunt T, Priestley A, Demengeot J, Mizuta R, Lehmann AR, Alt FW, Jackson SP, Jeggo PA. Ku80: product of the XRCC5 gene and its role in DNA repair and V(D)J recombination. Science. 1994;265:1442–1445. - PubMed
-
- Morio T, Kim H. Ku, Artemis, and ataxia-telangiectasia-mutated: signalling networks in DNA damage. Int. J. Biochem. Cell Biol. 2008;40:598–603. - PubMed
-
- Feldmann H, Driller L, Meier B, Mages G, Kellermann J, Winnacker EL. HDF2, the second subunit of the Ku homologue from Saccharomyces cerevisiae. J. Biol. Chem. 1996;271:27765–27769. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

