Strict control of transgene expression in a mouse model for sensitive biological applications based on RMCE compatible ES cells
- PMID: 20935052
- PMCID: PMC3017619
- DOI: 10.1093/nar/gkq868
Strict control of transgene expression in a mouse model for sensitive biological applications based on RMCE compatible ES cells
Abstract
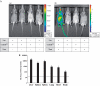
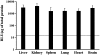
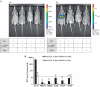
Recombinant mouse strains that harbor tightly controlled transgene expression proved to be indispensible tools to elucidate gene function. Different strategies have been employed to achieve controlled induction of the transgene. However, many models are accompanied by a considerable level of basal expression in the non-induced state. Thereby, applications that request tight control of transgene expression, such as the expression of toxic genes and the investigation of immune response to neo antigens are excluded. We developed a new Cre/loxP-based strategy to achieve strict control of transgene expression. This strategy was combined with RMCE (recombinase mediated cassette exchange) that facilitates the targeting of genes into a tagged site in ES cells. The tightness of regulation was confirmed using luciferase as a reporter. The transgene was induced upon breeding these mice to effector animals harboring either the ubiquitous (ROSA26) or liver-specific (Albumin) expression of CreER(T2), and subsequent feeding with Tamoxifen. Making use of RMCE, luciferase was replaced by Ovalbumin antigen. Mice generated from these ES cells were mated with mice expressing liver-specific CreER(T2). The transgenic mice were examined for the establishment of an immune response. They were fully competent to establish an immune response upon hepatocyte specific OVA antigen expression as indicated by a massive liver damage upon Tamoxifen treatment and did not show OVA tolerance. Together, this proves that this strategy supports strict control of transgenes that is even compatible with highly sensitive biological readouts.
Figures




References
-
- Branda CS, Dymecki SM. Talking about a revolution: the impact of site-specific recombinases on genetic analyses in mice. Dev Cell. 2004;6:7–28. - PubMed
-
- Gossen M, Bujard H. Studying gene function in eukaryotes by conditional gene inactivation. Annu. Rev. Genet. 2002;36:153–173. - PubMed
-
- Lewandoski M. Conditional control of gene expression in the mouse. Nat. Rev. Genet. 2001;2:743–755. - PubMed
-
- Sun Y, Chen X, Xiao D. Tetracycline-inducible expression systems: new strategies and practices in the transgenic mouse modeling. Acta Biochim Biophys Sin. 2007;39:235–246. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

