A novel functional DEC1 promoter polymorphism -249T>C reduces risk of squamous cell carcinoma of the head and neck
- PMID: 20935061
- PMCID: PMC2994282
- DOI: 10.1093/carcin/bgq198
A novel functional DEC1 promoter polymorphism -249T>C reduces risk of squamous cell carcinoma of the head and neck
Abstract
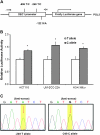
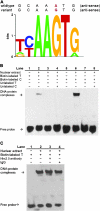
Human DEC1 (deleted in esophageal cancer 1) gene is located on chromosome 9q, a region frequently deleted in various types of human cancers, including squamous cell carcinoma of the head and neck (SCCHN). However, only one epidemiological study has evaluated the association between DEC1 polymorphisms and cancer risk. In this hospital-based case-control study, four potentially functional single-nucleotide polymorphisms -1628 G>A (rs1591420), -606 T>C [rs4978620, in complete linkage disequilibrium with -249T>C (rs2012775) and -122 G>A(rs2012566)], c.179 C>T p.Ala60Val (rs2269700) and 3' untranslated region-rs3750505 as well as the TP53 tumor suppressor gene codon 72 (Arg72Pro, rs1042522) polymorphism were genotyped in 1111 non-Hispanic Whites SCCHN patients and 1130 age-and sex-matched cancer-free controls. After adjustment for age, sex and smoking and drinking status, the variant -606CC (i.e. -249CC) homozygotes had a significantly reduced SCCHN risk (adjusted odds ratio = 0.71, 95% confidence interval = 0.52-0.99) compared with the -606TT homozygotes. Stratification analyses showed that a reduced risk associated with the -606CC genotype was more pronounced in subgroups of non-smokers, non-drinkers, younger subjects (defined as ≤57 years), carriers of the TP53 Arg/Arg (rs1042522) genotype, patients with oropharyngeal cancer or late-stage SCCHN. Further in silico analysis revealed that the -249 T-to-C change led to a gain of a transcription factor-binding site. Additional functional analysis showed that the -249T-to-C change significantly enhanced transcriptional activity of the DEC1 promoter and the DNA-protein-binding activity. We conclude that the DEC1 promoter -249 T>C (rs2012775) polymorphism is functional, modulating susceptibility to SCCHN among non-Hispanic Whites.
Figures
Similar articles
-
172G>T variant in the 5' untranslated region of DNA repair gene RAD51 reduces risk of squamous cell carcinoma of the head and neck and interacts with a P53 codon 72 variant.Carcinogenesis. 2007 May;28(5):988-94. doi: 10.1093/carcin/bgl225. Epub 2006 Nov 21. Carcinogenesis. 2007. PMID: 17118968
-
Single-nucleotide polymorphisms at the TP53-binding or responsive promoter regions of BAX and BCL2 genes and risk of squamous cell carcinoma of the head and neck.Carcinogenesis. 2007 Sep;28(9):2008-12. doi: 10.1093/carcin/bgm172. Epub 2007 Aug 11. Carcinogenesis. 2007. PMID: 17693666
-
Association of a p73 exon 2 G4C14-to-A4T14 polymorphism with risk of squamous cell carcinoma of the head and neck.Carcinogenesis. 2004 Oct;25(10):1911-6. doi: 10.1093/carcin/bgh197. Epub 2004 Jun 3. Carcinogenesis. 2004. PMID: 15180941
-
Effects of MDM2 promoter polymorphisms and p53 codon 72 polymorphism on risk and age at onset of squamous cell carcinoma of the head and neck.Mol Carcinog. 2011 Sep;50(9):697-706. doi: 10.1002/mc.20806. Epub 2011 Jun 7. Mol Carcinog. 2011. PMID: 21656578 Free PMC article.
-
Functional single nucleotide polymorphisms of the RASSF3 gene and susceptibility to squamous cell carcinoma of the head and neck.Eur J Cancer. 2014 Feb;50(3):582-92. doi: 10.1016/j.ejca.2013.11.009. Epub 2013 Nov 29. Eur J Cancer. 2014. PMID: 24295637 Free PMC article.
Cited by
-
Diversity of human clock genotypes and consequences.Prog Mol Biol Transl Sci. 2013;119:51-81. doi: 10.1016/B978-0-12-396971-2.00003-8. Prog Mol Biol Transl Sci. 2013. PMID: 23899594 Free PMC article. Review.
-
DNA Double-Strand Break Response and Repair Gene Polymorphisms May Influence Therapy Results and Prognosis in Head and Neck Cancer Patients.Cancers (Basel). 2023 Oct 13;15(20):4972. doi: 10.3390/cancers15204972. Cancers (Basel). 2023. PMID: 37894339 Free PMC article.
-
Molecular drivers of oral cavity squamous cell carcinoma in non-smoking and non-drinking patients: what do we know so far?Oncol Rev. 2022 Feb 22;16(1):549. doi: 10.4081/oncol.2022.549. eCollection 2022 Feb 22. Oncol Rev. 2022. PMID: 35340886 Free PMC article.
-
Functional variants of TIM-3/HAVCR2 3'UTR in lymphoblastoid cell lines.Future Sci OA. 2018 Mar 15;4(5):FSO298. doi: 10.4155/fsoa-2017-0121. eCollection 2018 Jun. Future Sci OA. 2018. PMID: 29796301 Free PMC article.
-
TP53 gene rs1042522 allele G decreases neuroblastoma risk: a two-centre case-control study.J Cancer. 2019 Jan 1;10(2):467-471. doi: 10.7150/jca.27482. eCollection 2019. J Cancer. 2019. PMID: 30719141 Free PMC article.
References
-
- Parkin DM, et al. Global cancer statistics, 2002. CA Cancer J. Clin. 2005;55:74–108. - PubMed
-
- Jemal A, et al. Cancer statistics, 2010. CA Cancer J. Clin. 2010;60:277–300. - PubMed
-
- Ho T, et al. Epidemiology of carcinogen metabolism genes and risk of squamous cell carcinoma of the head and neck. Head Neck. 2007;29:682–699. - PubMed
-
- Neumann AS, et al. Nucleotide excision repair as a marker for susceptibility to tobacco-related cancers: a review of molecular epidemiological studies. Mol. Carcinog. 2005;42:65–92. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

