Assembly and activation of a kinase ribozyme
- PMID: 20935068
- PMCID: PMC2995397
- DOI: 10.1261/rna.2302810
Assembly and activation of a kinase ribozyme
Abstract
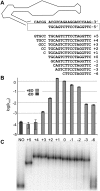
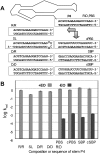
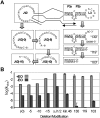
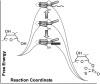
RNA activities can be regulated by modulating the relative energies of all conformations in a folding landscape; however, it is often unknown precisely how peripheral elements perturb the overall landscape in the absence of discrete alternative folds (inactive ensemble). This work explores the effects of sequence and secondary structure in governing kinase ribozyme activity. Kin.46 catalyzes thiophosphoryl transfer from ATPγS onto the 5' hydroxyl of polynucleotide substrates, and is regulated 10,000-fold by annealing an effector oligonucleotide to form activator helix P4. Transfer kinetics for an extensive series of ribozyme variants identified several dispensable internal single-stranded segments, in addition to a potential pseudoknot at the active site between segments J1/4 and J3/2 that is partially supported by compensatory rescue. Standard allosteric mechanisms were ruled out, such as formation of discrete repressive structures or docking P4 into the rest of the ribozyme via backbone 2' hydroxyls. Instead, P4 serves both to complete an important structural element (100-fold contribution to the reaction relative to a P4-deleted variant) and to mitigate nonspecific, inhibitory effects of the single-stranded tail (an additional 100-fold contribution to the apparent rate constant, k(obs)). Thermodynamic activation parameters ΔH(‡) and ΔS(‡), calculated from the temperature dependence of k(obs), varied with tail length and sequence. Inhibitory effects of the unpaired tail are largely enthalpic for short tails and are both enthalpic and entropic for longer tails. These results refine the structural view of this kinase ribozyme and highlight the importance of nonspecific ensemble effects in conformational regulation by peripheral elements.
Figures




References
-
- Achenbach J, Jeffries G, McManus S, Billen L, Li Y 2005. Secondary-structure characterization of two proficient kinase deoxyribozymes. Biochemistry 44: 3765–3774 - PubMed
-
- Admiraal S, Herschlag D 1995. Mapping the transition state for ATP hydrolysis: implications for enzymatic catalysis. Chem Biol 2: 729–739 - PubMed
-
- Admiraal S, Schneider B, Meyer P, Janin J, Véron M, Deville-Bonne D, Herschlag D 1999. Nucleophilic activation by positioning in phosphoryl transfer catalyzed by nucleoside diphosphate kinase. Biochemistry 38: 4701–4711 - PubMed
-
- Admiraal S, Meyer P, Schneider B, Deville-Bonne D, Janin J, Herschlag D 2001. Chemical rescue of phosphoryl transfer in a cavity mutant: A cautionary tale for site-directed mutagenesis. Biochemistry 40: 403–413 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
