Development of a sequence-characterized amplified region marker-targeted quantitative PCR assay for strain-specific detection of Oenococcus oeni during wine malolactic fermentation
- PMID: 20935116
- PMCID: PMC2988584
- DOI: 10.1128/AEM.00929-10
Development of a sequence-characterized amplified region marker-targeted quantitative PCR assay for strain-specific detection of Oenococcus oeni during wine malolactic fermentation
Abstract
Control over malolactic fermentation (MLF) is a difficult goal in winemaking and needs rapid methods to monitor Oenococcus oeni malolactic starters (MLS) in a stressful environment such as wine. In this study, we describe a novel quantitative PCR (QPCR) assay enabling the detection of an O. oeni strain during MLF without culturing. O. oeni strain LB221 was used as a model to develop a strain-specific sequence-characterized amplified region (SCAR) marker derived from a discriminatory OPA20-based randomly amplified polymorphic DNA (RAPD) band. The 5' and 3' flanking regions and the copy number of the SCAR marker were characterized using inverse PCR and Southern blotting, respectively. Primer pairs targeting the SCAR sequence enabled strain-specific detection without cross amplification of other O. oeni strains or wine species of lactic acid bacteria (LAB), acetic acid bacteria (AAB), and yeasts. The SCAR-QPCR assay was linear over a range of cell concentrations (7 log units) and detected as few as 2.2 × 10(2) CFU per ml of red wine with good quantification effectiveness, as shown by the correlation of QPCR and plate counting results. Therefore, the cultivation-independent monitoring of a single O. oeni strain in wine based on a SCAR marker represents a rapid and effective strain-specific approach. This strategy can be adopted to develop easy and rapid detection techniques for monitoring the implantation of inoculated O. oeni MLS on the indigenous LAB population, reducing the risk of unsuccessful MLF.
Figures
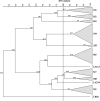




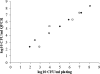

References
-
- Arnink, K., and T. Henick-Kling. 2005. Influence of Saccharomyces cerevisiae and Oenococcus oeni strains on successful malolactic conversion in wine. Am. J. Enol. Vitic. 56:228-237.
-
- Bartowsky, E. J., J. M. McCarthy, and P. Henschke. 2003. Differentiation of Australian wine isolates of Oenococcus oeni using random amplified polymorphic DNA (RAPD). Aust. J. Grape Wine Res. 9:122-126.
-
- Bouquet, R., J. Chirifie, and H. A. Iglesias. 1978. Equations for fitting water sorption isotherm of foods. II. Evaluation of various two-parameters models. J. Food Technol. 13:319-327.
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

