Derepression of INO1 transcription requires cooperation between the Ino2p-Ino4p heterodimer and Cbf1p and recruitment of the ISW2 chromatin-remodeling complex
- PMID: 20935143
- PMCID: PMC3008276
- DOI: 10.1128/EC.00144-10
Derepression of INO1 transcription requires cooperation between the Ino2p-Ino4p heterodimer and Cbf1p and recruitment of the ISW2 chromatin-remodeling complex
Abstract
The Saccharomyces cerevisiae INO1 gene encodes the structural enzyme inositol-3-phosphate synthase for the synthesis de novo of inositol and inositol-containing phospholipids. The transcription of INO1 is completely derepressed in the absence of inositol and choline (I(-) C(-)). Derepression requires the binding of the Ino2p-Ino4p basic helix-loop-helix (bHLH) heterodimer to the UAS(INO) promoter element. We report here the requirement of a third bHLH protein, centromere-binding factor 1 (Cbf1p), for the complete derepression of INO1 transcription. We found that Cbf1p regulates INO1 transcription by binding to sites distal to the INO1 promoter and encompassing the upstream SNA3 open reading frame (ORF) and promoter. The binding of Cbf1p requires Ino2p-Ino4p binding to the UAS(INO) sites in the INO1 promoter and vice versa, suggesting a cooperative mechanism. Furthermore, Cbf1p binding to the upstream sites was required for the binding of the ISW2 chromatin-remodeling complex to the Ino2p-Ino4p-binding sites on the INO1 promoter. Consistent with this, ISW2 was also required for the complete derepression of INO1 transcription.
Figures

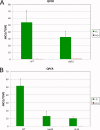
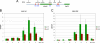

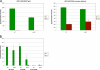


Similar articles
-
INO2 and INO4 gene products, positive regulators of phospholipid biosynthesis in Saccharomyces cerevisiae, form a complex that binds to the INO1 promoter.J Biol Chem. 1994 May 27;269(21):15344-9. J Biol Chem. 1994. PMID: 8195172
-
Gene-wide histone acetylation at the yeast INO1 requires the transcriptional activator Ino2p.Biochem Biophys Res Commun. 2010 Jan 8;391(2):1285-90. doi: 10.1016/j.bbrc.2009.12.063. Epub 2009 Dec 17. Biochem Biophys Res Commun. 2010. PMID: 20018175
-
Transcription regulation of the Saccharomyces cerevisiae PHO5 gene by the Ino2p and Ino4p basic helix-loop-helix proteins.Mol Microbiol. 2012 Jan;83(2):395-407. doi: 10.1111/j.1365-2958.2011.07941.x. Epub 2011 Dec 19. Mol Microbiol. 2012. PMID: 22182244
-
Structural Analysis of Ino2p/Ino4p Mutual Interactions and Their Binding Interface with Promoter DNA.Int J Mol Sci. 2022 Jul 9;23(14):7600. doi: 10.3390/ijms23147600. Int J Mol Sci. 2022. PMID: 35886947 Free PMC article.
-
ISWI complexes in Saccharomyces cerevisiae.Biochim Biophys Acta. 2004 Mar 15;1677(1-3):100-12. doi: 10.1016/j.bbaexp.2003.10.014. Biochim Biophys Acta. 2004. PMID: 15020051 Review.
Cited by
-
Rewiring phospholipid biosynthesis reveals resilience to membrane perturbations and uncovers regulators of lipid homeostasis.EMBO J. 2022 Apr 4;41(7):e109998. doi: 10.15252/embj.2021109998. Epub 2022 Feb 21. EMBO J. 2022. PMID: 35188676 Free PMC article.
-
Recruitment of an Activated Gene to the Yeast Nuclear Pore Complex Requires Sumoylation.Front Genet. 2020 Mar 6;11:174. doi: 10.3389/fgene.2020.00174. eCollection 2020. Front Genet. 2020. PMID: 32211027 Free PMC article.
-
Coordinated regulation of sulfur and phospholipid metabolism reflects the importance of methylation in the growth of yeast.Mol Biol Cell. 2011 Nov;22(21):4192-204. doi: 10.1091/mbc.E11-05-0467. Epub 2011 Sep 7. Mol Biol Cell. 2011. PMID: 21900497 Free PMC article.
-
Accumulation of unacetylatable Snf2p at the INO1 promoter is detrimental to remodeler recycling supply for CUP1 induction.PLoS One. 2020 Mar 25;15(3):e0230572. doi: 10.1371/journal.pone.0230572. eCollection 2020. PLoS One. 2020. PMID: 32210477 Free PMC article.
-
HLH-29 regulates ovulation in C. elegans by targeting genes in the inositol triphosphate signaling pathway.Biol Open. 2012 Mar 15;1(3):261-8. doi: 10.1242/bio.2012046. Epub 2012 Feb 8. Biol Open. 2012. PMID: 23213416 Free PMC article.
References
-
- Ambroziak J., Henry S. A. 1994. INO2 and INO4 gene products, positive regulators of phospholipid biosynthesis in Saccharomyces cerevisiae, form a complex that binds to the INO1 promoter. J. Biol. Chem. 269:15344–15349 - PubMed
-
- Aparicio O., Geisberg J. V., Struhl K. 2004. Chromatin immunoprecipitation for determining the association of proteins with specific genomic sequences in vivo. Curr. Protoc. Cell Biol. 17:17.17 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

