Essential role of glutathione in acclimation to environmental and redox perturbations in the cyanobacterium Synechocystis sp. PCC 6803
- PMID: 20935175
- PMCID: PMC2996012
- DOI: 10.1104/pp.110.162990
Essential role of glutathione in acclimation to environmental and redox perturbations in the cyanobacterium Synechocystis sp. PCC 6803
Abstract
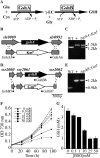
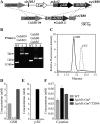
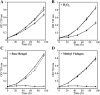
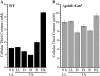
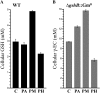
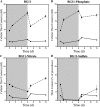
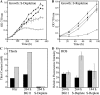
Glutathione, a nonribosomal thiol tripeptide, has been shown to be critical for many processes in plants. Much less is known about the roles of glutathione in cyanobacteria, oxygenic photosynthetic prokaryotes that are the evolutionary precursor of the chloroplast. An understanding of glutathione metabolism in cyanobacteria is expected to provide novel insight into the evolution of the elaborate and extensive pathways that utilize glutathione in photosynthetic organisms. To investigate the function of glutathione in cyanobacteria, we generated deletion mutants of glutamate-cysteine ligase (gshA) and glutathione synthetase (gshB) in Synechocystis sp. PCC 6803. Complete segregation of the ΔgshA mutation was not achieved, suggesting that GshA activity is essential for growth. In contrast, fully segregated ΔgshB mutants were isolated and characterized. The ΔgshB strain lacks reduced glutathione (GSH) but instead accumulates the precursor compound γ-glutamylcysteine (γ-EC). The ΔgshB strain grows slower than the wild-type strain under favorable conditions and exhibits extremely reduced growth or death when subjected to conditions promoting oxidative stress. Furthermore, we analyzed thiol contents in the wild type and the ΔgshB mutant after subjecting the strains to multiple environmental and redox perturbations. We found that conditions promoting growth stimulate glutathione biosynthesis. We also determined that cellular GSH and γ-EC content decline following exposure to dark and blue light and during photoheterotrophic growth. Moreover, a rapid depletion of GSH and γ-EC is observed in the wild type and the ΔgshB strain, respectively, when cells are starved for nitrate or sulfate.
Figures
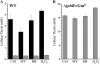

Similar articles
-
Glutathione in Synechocystis 6803: a closer look into the physiology of a ∆gshB mutant.Plant Signal Behav. 2011 Jan;6(1):89-92. doi: 10.4161/psb.6.1.14145. Epub 2011 Jan 1. Plant Signal Behav. 2011. PMID: 21301218 Free PMC article.
-
Probing the origins of glutathione biosynthesis through biochemical analysis of glutamate-cysteine ligase and glutathione synthetase from a model photosynthetic prokaryote.Biochem J. 2013 Feb 15;450(1):63-72. doi: 10.1042/BJ20121332. Biochem J. 2013. PMID: 23170977
-
First evidence of glutathione metabolism in Leptospira interrogans.Free Radic Biol Med. 2019 Nov 1;143:366-374. doi: 10.1016/j.freeradbiomed.2019.08.028. Epub 2019 Aug 26. Free Radic Biol Med. 2019. PMID: 31465831
-
Oxidative stress and regulation of glutathione in lung inflammation.Eur Respir J. 2000 Sep;16(3):534-54. doi: 10.1034/j.1399-3003.2000.016003534.x. Eur Respir J. 2000. PMID: 11028671 Review.
-
Glutathione: an overview of biosynthesis and modulation.Chem Biol Interact. 1998 Apr 24;111-112:1-14. doi: 10.1016/s0009-2797(97)00146-4. Chem Biol Interact. 1998. PMID: 9679538 Review.
Cited by
-
Comparative Genomics of Synechococcus elongatus Explains the Phenotypic Diversity of the Strains.mBio. 2022 Jun 28;13(3):e0086222. doi: 10.1128/mbio.00862-22. Epub 2022 Apr 27. mBio. 2022. PMID: 35475644 Free PMC article.
-
Adaptive Engineering of Phytochelatin-based Heavy Metal Tolerance.J Biol Chem. 2015 Jul 10;290(28):17321-30. doi: 10.1074/jbc.M115.652123. Epub 2015 May 27. J Biol Chem. 2015. PMID: 26018077 Free PMC article.
-
Eukaryotic and Prokaryotic Phytochelatin Synthases Differ Less in Functional Terms Than Previously Thought: A Comparative Analysis of Marchantia polymorpha and Geitlerinema sp. PCC 7407.Plants (Basel). 2020 Jul 20;9(7):914. doi: 10.3390/plants9070914. Plants (Basel). 2020. PMID: 32698350 Free PMC article.
-
Dark accumulation of downstream glycolytic intermediates initiates robust photosynthesis in cyanobacteria.Plant Physiol. 2023 Apr 3;191(4):2400-2413. doi: 10.1093/plphys/kiac602. Plant Physiol. 2023. PMID: 36574371 Free PMC article.
-
Urea Is Both a Carbon and Nitrogen Source for Microcystis aeruginosa: Tracking 13C Incorporation at Bloom pH Conditions.Front Microbiol. 2019 May 17;10:1064. doi: 10.3389/fmicb.2019.01064. eCollection 2019. Front Microbiol. 2019. PMID: 31164875 Free PMC article.
References
-
- Allen MM. (1968) Simple conditions for growth of unicellular blue-green algae on plates. J Phycol 4: 1–4 - PubMed
-
- Asada K. (1999) The water-water cycle in chloroplasts: scavenging of active oxygens and dissipation of excess photons. Annu Rev Plant Physiol Plant Mol Biol 50: 601–639 - PubMed
-
- Ashida H, Sawa Y, Shibata H. (2005) Cloning, biochemical and phylogenetic characterizations of gamma-glutamylcysteine synthetase from Anabaena sp. PCC 7120. Plant Cell Physiol 46: 557–562 - PubMed
-
- Aurora R, Hihara Y, Singh AK, Pakrasi HB. (2007) A network of genes regulated by light in cyanobacteria. OMICS 11: 166–185 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

