MGMT-independent temozolomide resistance in pediatric glioblastoma cells associated with a PI3-kinase-mediated HOX/stem cell gene signature
- PMID: 20935218
- PMCID: PMC3935452
- DOI: 10.1158/0008-5472.CAN-10-1250
MGMT-independent temozolomide resistance in pediatric glioblastoma cells associated with a PI3-kinase-mediated HOX/stem cell gene signature
Abstract
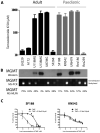
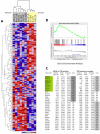
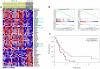
Sensitivity to temozolomide is restricted to a subset of glioblastoma patients, with the major determinant of resistance being a lack of promoter methylation of the gene encoding the repair protein DNA methyltransferase MGMT, although other mechanisms are thought to be active. There are, however, limited preclinical data in model systems derived from pediatric glioma patients. We screened a series of cell lines for temozolomide efficacy in vitro, and investigated the differential mechanisms of resistance involved. In the majority of cell lines, a lack of MGMT promoter methylation and subsequent protein overexpression were linked to temozolomide resistance. An exception was the pediatric glioblastoma line KNS42. Expression profiling data revealed a coordinated upregulation of HOX gene expression in resistant lines, especially KNS42, which was reversed by phosphoinositide 3-kinase pathway inhibition. High levels of HOXA9/HOXA10 gene expression were associated with a shorter survival in pediatric high-grade glioma patient samples. Combination treatment in vitro of pathway inhibition and temozolomide resulted in a highly synergistic interaction in KNS42 cells. The resistance gene signature further included contiguous genes within the 12q13-q14 amplicon, including the Akt enhancer PIKE, significantly overexpressed in the KNS42 line. These cells were also highly enriched for CD133 and other stem cell markers. We have thus shown an in vitro link between phosphoinositide 3-kinase-mediated HOXA9/HOXA10 expression, and a drug-resistant, progenitor cell phenotype in MGMT-independent pediatric glioblastoma.
Copyright © 2010 AACR.
Figures



References
-
- Mrugala MM, Chamberlain MC. Mechanisms of disease: temozolomide and glioblastoma--look to the future. Nat Clin Pract Oncol. 2008;5:476–86. - PubMed
-
- Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–96. - PubMed
-
- Hegi ME, Diserens AC, Gorlia T, Hamou MF, de Tribolet N, Weller M, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. - PubMed
-
- Esteller M, Garcia-Foncillas J, Andion E, Goodman SN, Hidalgo OF, Vanaclocha V, et al. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med. 2000;343:1350–4. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

