Mitochondrial beta-cyanoalanine synthase is essential for root hair formation in Arabidopsis thaliana
- PMID: 20935247
- PMCID: PMC2990132
- DOI: 10.1105/tpc.110.076828
Mitochondrial beta-cyanoalanine synthase is essential for root hair formation in Arabidopsis thaliana
Abstract
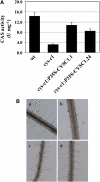
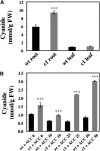
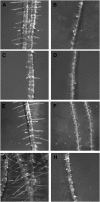
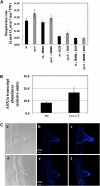
Cyanide is stoichiometrically produced as a coproduct of the ethylene biosynthesis pathway and is detoxified by β-cyanoalanine synthase enzymes. The molecular and phenotypical analysis of T-DNA insertion mutants of the mitochondrial β-cyanoalanine synthase CYS-C1 suggests that discrete accumulation of cyanide is not toxic for the plant and does not alter mitochondrial respiration rates but does act as a strong inhibitor of root hair development. The cys-c1 null allele is defective in root hair formation and accumulates cyanide in root tissues. The root hair defect is phenocopied in wild-type plants by the exogenous addition of cyanide to the growth medium and is reversed by the addition of hydroxocobalamin or by genetic complementation with the CYS-C1 gene. Hydroxocobalamin not only recovers the root phenotype of the mutant but also the formation of reactive oxygen species at the initial step of root hair tip growth. Transcriptional profiling of the cys-c1 mutant reveals that cyanide accumulation acts as a repressive signal for several genes encoding enzymes involved in cell wall rebuilding and the formation of the root hair tip as well as genes involved in ethylene signaling and metabolism. Our results demonstrate that mitochondrial β-cyanoalanine synthase activity is essential to maintain a low level of cyanide for proper root hair development.
Figures



Similar articles
-
ß-Cyanoalanine Synthase Action in Root Hair Elongation is Exerted at Early Steps of the Root Hair Elongation Pathway and is Independent of Direct Cyanide Inactivation of NADPH Oxidase.Plant Cell Physiol. 2018 May 1;59(5):1072-1083. doi: 10.1093/pcp/pcy047. Plant Cell Physiol. 2018. PMID: 29490083
-
Transient transcriptional regulation of the CYS-C1 gene and cyanide accumulation upon pathogen infection in the plant immune response.Plant Physiol. 2013 Aug;162(4):2015-27. doi: 10.1104/pp.113.219436. Epub 2013 Jun 19. Plant Physiol. 2013. PMID: 23784464 Free PMC article.
-
Beyond toxicity: a regulatory role for mitochondrial cyanide.Plant Signal Behav. 2014;9(1):e27612. doi: 10.4161/psb.27612. Epub 2014 Jan 7. Plant Signal Behav. 2014. PMID: 24398435 Free PMC article.
-
The β-cyanoalanine synthase pathway: beyond cyanide detoxification.Plant Cell Environ. 2016 Oct;39(10):2329-41. doi: 10.1111/pce.12755. Epub 2016 Jun 16. Plant Cell Environ. 2016. PMID: 27116378 Review.
-
Signaling by hydrogen sulfide and cyanide through post-translational modification.J Exp Bot. 2019 Aug 19;70(16):4251-4265. doi: 10.1093/jxb/erz225. J Exp Bot. 2019. PMID: 31087094 Review.
Cited by
-
Role of mitochondrial cyanide detoxification in Arabidopsis root hair development.Plant Signal Behav. 2018;13(12):e1537699. doi: 10.1080/15592324.2018.1537699. Epub 2018 Oct 31. Plant Signal Behav. 2018. PMID: 30380363 Free PMC article.
-
Genome-Wide Investigation of the Cysteine Synthase Gene Family Shows That Overexpression of CSase Confers Alkali Tolerance to Alfalfa (Medicago sativa L.).Front Plant Sci. 2022 Jan 4;12:792862. doi: 10.3389/fpls.2021.792862. eCollection 2021. Front Plant Sci. 2022. PMID: 35058952 Free PMC article.
-
A Cyanide-Induced 3-Cyanoalanine Nitrilase in the Cyanide-Assimilating Bacterium Pseudomonas pseudoalcaligenes Strain CECT 5344.Appl Environ Microbiol. 2017 Apr 17;83(9):e00089-17. doi: 10.1128/AEM.00089-17. Print 2017 May 1. Appl Environ Microbiol. 2017. PMID: 28235872 Free PMC article.
-
Structure of soybean β-cyanoalanine synthase and the molecular basis for cyanide detoxification in plants.Plant Cell. 2012 Jun;24(6):2696-706. doi: 10.1105/tpc.112.098954. Epub 2012 Jun 26. Plant Cell. 2012. PMID: 22739827 Free PMC article.
-
Negative Regulation of Autophagy by Sulfide Is Independent of Reactive Oxygen Species.Plant Physiol. 2016 Jun;171(2):1378-91. doi: 10.1104/pp.16.00110. Epub 2016 Apr 14. Plant Physiol. 2016. PMID: 27208225 Free PMC article.
References
-
- Albury M.S., Elliott C., Moore A.L. (2009). Towards a structural elucidation of the alternative oxidase in plants. Physiol. Plant. 137: 316–327 - PubMed
-
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 - PubMed
-
- Astier A., Baud F.J. (1996). Complexation of intracellular cyanide by hydroxocobalamin using a human cellular model. Hum. Exp. Toxicol. 15: 19–25 - PubMed
-
- Babula D., Misztal L.H., Jakubowicz M., Kaczmarek M., Nowak W., Sadowski J. (2006). Genes involved in biosynthesis and signalisation of ethylene in Brassica oleracea and Arabidopsis thaliana: Identification and genome comparative mapping of specific gene homologues. Theor. Appl. Genet. 112: 410–420 - PubMed
-
- Bariola P.A., Howard C.J., Taylor C.B., Verburg M.T., Jaglan V.D., Green P.J. (1994). The Arabidopsis ribonuclease gene RNS1 is tightly controlled in response to phosphate limitation. Plant J. 6: 673–685 - PubMed
MeSH terms
Substances
Associated data
- Actions

