Rmi1 stimulates decatenation of double Holliday junctions during dissolution by Sgs1-Top3
- PMID: 20935631
- PMCID: PMC2988882
- DOI: 10.1038/nsmb.1919
Rmi1 stimulates decatenation of double Holliday junctions during dissolution by Sgs1-Top3
Abstract
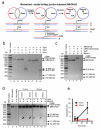
A double Holliday junction (dHJ) is a central intermediate of homologous recombination that can be processed to yield crossover or non-crossover recombination products. To preserve genomic integrity, cells possess mechanisms to avoid crossing over. We show that Saccharomyces cerevisiae Sgs1 and Top3 proteins are sufficient to migrate and disentangle a dHJ to produce exclusively non-crossover recombination products, in a reaction termed "dissolution." We show that Rmi1 stimulates dHJ dissolution at low Sgs1-Top3 protein concentrations, although it has no effect on the initial rate of Holliday junction (HJ) migration. Rmi1 serves to stimulate DNA decatenation, removing the last linkages between the repaired and template DNA molecules. Dissolution of a dHJ is a highly efficient and concerted alternative to nucleolytic resolution that prevents crossing over of chromosomes during recombinational DNA repair in mitotic cells and thereby contributes to genomic integrity.
Figures
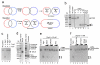



References
-
- Klein HL, Symington LS. Breaking up just got easier to do. Cell. 2009;138:20–2. - PubMed
-
- Heyer WD, Ehmsen KT, Solinger JA. Holliday junctions in the eukaryotic nucleus: resolution in sight? Trends Biochem Sci. 2003;28:548–57. - PubMed
-
- Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nat Rev Mol Cell Biol. 2002;3:430–440. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

