PCRless library mutagenesis via oligonucleotide recombination in yeast
- PMID: 20936671
- PMCID: PMC3009401
- DOI: 10.1002/pro.513
PCRless library mutagenesis via oligonucleotide recombination in yeast
Abstract
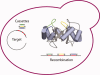
The directed evolution of biomolecules with new functions is largely performed in vitro, with PCR mutagenesis followed by high-throughput assays for desired activities. As synthetic biology creates impetus for generating biomolecules that function in living cells, new technologies are needed for performing mutagenesis and selection for directed evolution in vivo. Homologous recombination, routinely exploited for targeted gene alteration, is an attractive tool for in vivo library mutagenesis, yet surprisingly is not routinely used for this purpose. Here, we report the design and characterization of a yeast-based system for library mutagenesis of protein loops via oligonucleotide recombination. In this system, a linear vector is co-transformed with single-stranded mutagenic oligonucleotides. Using repair of nonsense codons engineered in three different active-site loops in the selectable marker TRP1 as a model system, we first optimized the recombination efficiency. Single-loop recombination was highly efficient, averaging 5%, or 4.0×10(5) recombinants. Multiple loops could be simultaneously mutagenized, although the efficiencies dropped to 0.2%, or 6.0×10(3) recombinants, for two loops and 0.01% efficiency, or 1.5×10(2) recombinants, for three loops. Finally, the utility of this system for directed evolution was tested explicitly by selecting functional variants from a mock library of 1:10(6) wild-type:nonsense codons. Sequencing showed that oligonucleotide recombination readily covered this large library, mutating not only the target codon but also encoded silent mutations on either side of the library cassette. Together these results establish oligonucleotide recombination as a simple and powerful library mutagenesis technique and advance efforts to engineer the cell for fully in vivo directed evolution.
Copyright © 2010 The Protein Society.
Figures
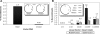


References
-
- Boder ET, Wittrup KD. Yeast surface display for screening combinatorial polypeptide libraries. Nat Biotechnol. 1997;15:553–557. - PubMed
-
- Smith M. In vitro mutagenesis. Annu Rev Genet. 1985;19:423–462. - PubMed
-
- Bloom JD, Meyer MM, Meinhold P, Otey CR, MacMillan D, Arnold FH. Evolving strategies for enzyme engineering. Curr Opin Struct Biol. 2005;15:447–452. - PubMed
-
- Botstein D, Shortle D. Strategies and applications of in vitro mutagenesis. Science. 1985;229:1193–1201. - PubMed
-
- Klinner U, Schafer B. Genetic aspects of targeted insertion mutagenesis in yeasts. FEMS Microbiol Rev. 2004;28:201–223. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

