RAD18-mediated ubiquitination of PCNA activates the Fanconi anemia DNA repair network
- PMID: 20937699
- PMCID: PMC2958487
- DOI: 10.1083/jcb.201005101
RAD18-mediated ubiquitination of PCNA activates the Fanconi anemia DNA repair network
Abstract
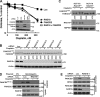
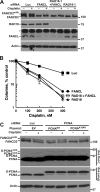
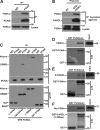
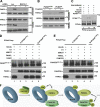
The Fanconi anemia (FA) network is important for the repair of interstrand DNA cross-links. A key event in FA pathway activation is the monoubiquitylation of the FA complementation group I (FANCI)-FANCD2 (ID) complex by FA complementation group L (FANCL), an E3 ubiquitin ligase. In this study, we show that RAD18, another DNA damage-activated E3 ubiquitin ligase, also participates in ID complex activation by ubiquitylating proliferating cell nuclear antigen (PCNA) on Lys164, an event required for the recruitment of FANCL to chromatin. We also found that monoubiquitylated PCNA stimulates FANCL-catalyzed FANCD2 and FANCI monoubiquitylation. Collectively, these experiments identify RAD18-mediated PCNA monoubiquitination as a central hub for the mobilization of the FA pathway by promoting FANCL-mediated FANCD2 monoubiquitylation.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

