Family-wide characterization of the DENN domain Rab GDP-GTP exchange factors
- PMID: 20937701
- PMCID: PMC2958468
- DOI: 10.1083/jcb.201008051
Family-wide characterization of the DENN domain Rab GDP-GTP exchange factors
Abstract
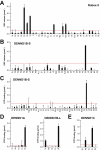
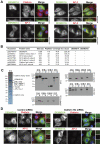
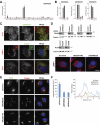
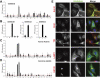
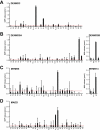
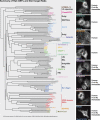
A key requirement for Rab function in membrane trafficking is site-specific activation by GDP-GTP exchange factors (GEFs), but the majority of the 63 human Rabs have no known GEF. We have performed a systematic characterization of the 17 human DENN domain proteins and demonstrated that they are specific GEFs for 10 Rabs. DENND1A/1B localize to clathrin patches at the plasma membrane and activate Rab35 in an endocytic pathway trafficking Shiga toxin to the trans-Golgi network. DENND2 GEFs target to actin filaments and control Rab9-dependent trafficking of mannose-6-phosphate receptor to lysosomes. DENND4 GEFs target to a tubular membrane compartment adjacent to the Golgi, where they activate Rab10, which suggests a function in basolateral polarized sorting in epithelial cells that compliments the non-DENN GEF Sec2 acting on Rab8 in apical sorting. DENND1C, DENND3, DENND5A/5B, MTMR5/13, and MADD activate Rab13, Rab12, Rab39, Rab28, and Rab27A/27B, respectively. Together, these findings provide a basis for future studies on Rab regulation and function.
Figures


References
-
- Azzedine H., Bolino A., Taïeb T., Birouk N., Di Duca M., Bouhouche A., Benamou S., Mrabet A., Hammadouche T., Chkili T., et al. 2003. Mutations in MTMR13, a new pseudophosphatase homologue of MTMR2 and Sbf1, in two families with an autosomal recessive demyelinating form of Charcot-Marie-Tooth disease associated with early-onset glaucoma. Am. J. Hum. Genet. 72:1141–1153 10.1086/375034 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

