Drosophila Myc interacts with host cell factor (dHCF) to activate transcription and control growth
- PMID: 20937797
- PMCID: PMC3000943
- DOI: 10.1074/jbc.M110.140467
Drosophila Myc interacts with host cell factor (dHCF) to activate transcription and control growth
Abstract
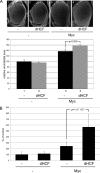
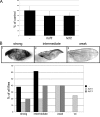
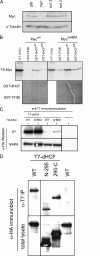
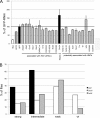
The Myc proto-oncoproteins are transcription factors that recognize numerous target genes through hexameric DNA sequences called E-boxes. The mechanism by which they then activate the expression of these targets is still under debate. Here, we use an RNAi screen in Drosophila S2 cells to identify Drosophila host cell factor (dHCF) as a novel co-factor for Myc that is functionally required for the activation of a Myc-dependent reporter construct. dHCF is also essential for the full activation of endogenous Myc target genes in S2 cells, and for the ability of Myc to promote growth in vivo. Myc and dHCF physically interact, and they colocalize on common target genes. Furthermore, down-regulation of dHCF-associated histone acetyltransferase and histone methyltransferase complexes in vivo interferes with the Myc biological activities. We therefore propose that dHCF recruits such chromatin-modifying complexes and thereby contributes to the expression of Myc targets and hence to the execution of Myc biological activities.
Figures



Similar articles
-
Histone H3 lysine 4 trimethylation regulates cotranscriptional H2A variant exchange by Tip60 complexes to maximize gene expression.Proc Natl Acad Sci U S A. 2014 Apr 1;111(13):4850-5. doi: 10.1073/pnas.1320337111. Epub 2014 Mar 17. Proc Natl Acad Sci U S A. 2014. PMID: 24639513 Free PMC article.
-
Drosophila melanogaster dHCF interacts with both PcG and TrxG epigenetic regulators.PLoS One. 2011;6(12):e27479. doi: 10.1371/journal.pone.0027479. Epub 2011 Dec 8. PLoS One. 2011. PMID: 22174740 Free PMC article.
-
The Trithorax group protein Lid is a trimethyl histone H3K4 demethylase required for dMyc-induced cell growth.Genes Dev. 2007 Mar 1;21(5):537-51. doi: 10.1101/gad.1523007. Epub 2007 Feb 20. Genes Dev. 2007. PMID: 17311883 Free PMC article.
-
Chromatin mechanisms in Drosophila dosage compensation.Prog Mol Subcell Biol. 2005;38:123-49. doi: 10.1007/3-540-27310-7_5. Prog Mol Subcell Biol. 2005. PMID: 15881893 Review.
-
RNA at the steering wheel.Genome Biol. 2006;7(5):218. doi: 10.1186/gb-2006-7-5-218. Epub 2006 May 26. Genome Biol. 2006. PMID: 16732899 Free PMC article. Review.
Cited by
-
PAF1 complex component Leo1 helps recruit Drosophila Myc to promoters.Proc Natl Acad Sci U S A. 2017 Oct 31;114(44):E9224-E9232. doi: 10.1073/pnas.1705816114. Epub 2017 Oct 16. Proc Natl Acad Sci U S A. 2017. PMID: 29078288 Free PMC article.
-
Myc function in Drosophila.Cold Spring Harb Perspect Med. 2013 Oct 1;3(10):a014324. doi: 10.1101/cshperspect.a014324. Cold Spring Harb Perspect Med. 2013. PMID: 24086064 Free PMC article. Review.
-
Spatiotemporal control of cell ablation using Ronidazole with Nitroreductase in Drosophila.Dev Biol. 2025 Apr;520:31-40. doi: 10.1016/j.ydbio.2024.12.017. Epub 2024 Dec 28. Dev Biol. 2025. PMID: 39736378
-
Evidence for tissue-specific Jak/STAT target genes in Drosophila optic lobe development.Genetics. 2013 Dec;195(4):1291-306. doi: 10.1534/genetics.113.155945. Epub 2013 Sep 27. Genetics. 2013. PMID: 24077308 Free PMC article.
-
Ribogenesis boosts controlled by HEATR1-MYC interplay promote transition into brain tumour growth.EMBO Rep. 2024 Jan;25(1):168-197. doi: 10.1038/s44319-023-00017-1. Epub 2024 Jan 15. EMBO Rep. 2024. PMID: 38225354 Free PMC article.
References
-
- Oster S. K., Ho C. S., Soucie E. L., Penn L. Z. (2002) Adv. Cancer Res. 84, 81–154 - PubMed
-
- Pirity M., Blanck J. K., Schreiber-Agus N. (2006) Curr. Top Microbiol. Immunol. 302, 205–234 - PubMed
-
- Trumpp A., Refaeli Y., Oskarsson T., Gasser S., Murphy M., Martin G. R., Bishop J. M. (2001) Nature 414, 768–773 - PubMed
-
- Gallant P. (2009) Adv. Cancer Res. 103, 111–144 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

