A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage
- PMID: 20937877
- PMCID: PMC2972950
- DOI: 10.1073/pnas.1012946107
A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage
Abstract
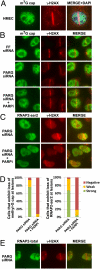
Many proteins that respond to DNA damage are recruited to DNA lesions. We used a proteomics approach that coupled isotopic labeling with chromatin fractionation and mass spectrometry to uncover proteins that associate with damaged DNA, many of which are involved in DNA repair or nucleolar function. We show that polycomb group members are recruited by poly(ADP ribose) polymerase (PARP) to DNA lesions following UV laser microirradiation. Loss of polycomb components results in IR sensitivity of mammalian cells and Caenorhabditis elegans. PARP also recruits two components of the repressive nucleosome remodeling and deacetylase (NuRD) complex, chromodomain helicase DNA-binding protein 4 (CHD4) and metastasis associated 1 (MTA1), to DNA lesions. PARP plays a role in removing nascent RNA and elongating RNA polymerase II from sites of DNA damage. We propose that PARP sets up a transient repressive chromatin structure at sites of DNA damage to block transcription and facilitate DNA repair.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

