A novel heme-responsive element mediates transcriptional regulation in Caenorhabditis elegans
- PMID: 20938051
- PMCID: PMC2998115
- DOI: 10.1074/jbc.M110.167619
A novel heme-responsive element mediates transcriptional regulation in Caenorhabditis elegans
Abstract
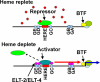
Hemes are prosthetic groups that participate in diverse biochemical pathways across phylogeny. Although heme can also regulate broad physiological processes by directly modulating gene expression in Metazoa, the regulatory pathways for sensing and responding to heme are not well defined. Caenorhabditis elegans is a heme auxotroph and relies solely on environmental heme for sustenance. Worms respond to heme availability by regulating heme-responsive genes such as hrg-1, an intestinal heme transporter that is up-regulated by >60-fold during heme depletion. To identify the mechanism for the heme-dependent regulation of hrg-1, we interrogated the hrg-1 promoter. Deletion and mutagenesis studies of the hrg-1 promoter revealed a 23-bp heme-responsive element that is both necessary and sufficient for heme-dependent regulation of hrg-1. Furthermore, our studies show that the heme regulation of hrg-1 is mediated by both activation and repression in conjunction with ELT-2 and ELT-4, transcription factors that specify intestinal expression.
Figures






References
-
- Ajioka R. S., Phillips J. D., Kushner J. P. (2006) Biochim. Biophys. Acta 1763, 723–736 - PubMed
-
- Rodgers K. R. (1999) Curr. Opin. Chem. Biol. 3, 158–167 - PubMed
-
- Wenger R. H. (2000) J. Exp. Biol. 203, 1253–1263 - PubMed
-
- Gilles-Gonzalez M. A., Gonzalez G. (2004) J. Appl. Physiol. 96, 774–783 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

