Subtype-associated differences in HIV-1 reverse transcription affect the viral replication
- PMID: 20939905
- PMCID: PMC2964588
- DOI: 10.1186/1742-4690-7-85
Subtype-associated differences in HIV-1 reverse transcription affect the viral replication
Abstract
Background: The impact of the products of the pol gene, specifically, reverse transcriptase (RT) on HIV-1 replication, evolution, and acquisition of drug resistance has been thoroughly characterized for subtype B. For subtype C, which accounts of almost 60% of HIV cases worldwide, much less is known. It has been reported that subtype C HIV-1 isolates have a lower replication capacity than B; however, the basis of these differences remains unclear.
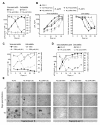
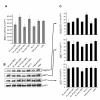
Results: We analyzed the impact of the pol gene products from HIV-1 B and C subtypes on the maturation of HIV virions, accumulation of reverse transcription products, integration of viral DNA, frequency of point mutations in provirus and overall viral replication. Recombinant HIV-1 viruses of B and C subtypes comprising the pol fragments encoding protease, integrase and either the whole RT or a chimeric RT from different isolates of the C and B subtypes, were used for infection of cells expressing CXCR4 or CCR5 co-receptors. The viruses carrying different fragments of pol from the isolates of B and C subtypes did not reveal differences in Gag and GagPol processing and viral RNA incorporation into the virions. However, the presence of the whole RT from subtype C, or the chimeric RT containing either the polymerase or the connection and RNase H domains from C isolates, caused significantly slower viral replication regardless of B or C viral backbone. Subtype C RT carrying viruses displayed lower levels of accumulation of strong-stop cDNA in permeabilized virions during endogenous reverse transcription, and decreased accumulation of both strong-stop and positive strand reverse transcription products in infected cells and in isolated reverse transcription complexes. This decreased accumulation correlated with lower levels of viral DNA integration in cells infected with viruses carrying the whole RT or RT domains from subtype C isolates. The single viral genome assay analysis did not reveal significant differences in the frequency of point mutations between the RT from B or C subtypes.
Conclusions: These data suggest that the whole RT as well as distinct polymerase and connection-RNase H domains from subtype C HIV-1 confer a lower level of accumulation of reverse transcripts in the virions and reverse transcription complexes as compared to subtype B, resulting in a lower overall level of virus replication.
Figures



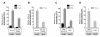
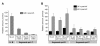
Similar articles
-
Pol-Driven Replicative Capacity Impacts Disease Progression in HIV-1 Subtype C Infection.J Virol. 2018 Sep 12;92(19):e00811-18. doi: 10.1128/JVI.00811-18. Print 2018 Oct 1. J Virol. 2018. PMID: 29997209 Free PMC article.
-
Virion instability of human immunodeficiency virus type 1 reverse transcriptase (RT) mutated in the protease cleavage site between RT p51 and the RT RNase H domain.J Virol. 2005 Sep;79(18):11952-61. doi: 10.1128/JVI.79.18.11952-11961.2005. J Virol. 2005. PMID: 16140771 Free PMC article.
-
Subtype-Specific Differences in Gag-Protease-Driven Replication Capacity Are Consistent with Intersubtype Differences in HIV-1 Disease Progression.J Virol. 2017 Jun 9;91(13):e00253-17. doi: 10.1128/JVI.00253-17. Print 2017 Jul 1. J Virol. 2017. PMID: 28424286 Free PMC article.
-
Cutting into the Substrate Dominance: Pharmacophore and Structure-Based Approaches toward Inhibiting Human Immunodeficiency Virus Reverse Transcriptase-Associated Ribonuclease H.Acc Chem Res. 2020 Jan 21;53(1):218-230. doi: 10.1021/acs.accounts.9b00450. Epub 2019 Dec 27. Acc Chem Res. 2020. PMID: 31880912 Free PMC article. Review.
-
Polymorphism in HIV-1 non-subtype B protease and reverse transcriptase and its potential impact on drug susceptibility and drug resistance evolution.AIDS Rev. 2003 Jan-Mar;5(1):25-35. AIDS Rev. 2003. PMID: 12875105 Review.
Cited by
-
Evolutionary Analysis of HIV-1 Pol Proteins Reveals Representative Residues for Viral Subtype Differentiation.Front Microbiol. 2017 Nov 2;8:2151. doi: 10.3389/fmicb.2017.02151. eCollection 2017. Front Microbiol. 2017. PMID: 29163435 Free PMC article.
-
Consequences of HIV infection in the bone marrow niche.Front Immunol. 2023 Jul 11;14:1163012. doi: 10.3389/fimmu.2023.1163012. eCollection 2023. Front Immunol. 2023. PMID: 37497228 Free PMC article. Review.
-
Differential Vpu-Mediated CD4 and Tetherin Downregulation Functions among Major HIV-1 Group M Subtypes.J Virol. 2020 Jul 1;94(14):e00293-20. doi: 10.1128/JVI.00293-20. Print 2020 Jul 1. J Virol. 2020. PMID: 32376625 Free PMC article.
-
HIV-1 Latency and Latency Reversal: Does Subtype Matter?Viruses. 2019 Nov 28;11(12):1104. doi: 10.3390/v11121104. Viruses. 2019. PMID: 31795223 Free PMC article. Review.
-
Ability of HIV-1 Nef to downregulate CD4 and HLA class I differs among viral subtypes.Retrovirology. 2013 Sep 16;10:100. doi: 10.1186/1742-4690-10-100. Retrovirology. 2013. PMID: 24041011 Free PMC article.
References
-
- de Oliveira T, Engelbrecht S, Janse van Rensburg E, Gordon M, Bishop K, zur Megede J, Barnett SW, Cassol S. Variability at human immunodeficiency virus type 1 subtype C protease cleavage sites: an indication of viral fitness? J Virol. 2003;77:9422–9430. doi: 10.1128/JVI.77.17.9422-9430.2003. - DOI - PMC - PubMed
-
- Korber BT, Allen EE, Farmer AD, Myers GL. Heterogeneity of HIV-1 and HIV-2. AIDS. 1995;9(Suppl A):S5–18. - PubMed
-
- Loemba H, Brenner B, Parniak MA, Ma'ayan S, Spira B, Moisi D, Oliveira M, Detorio M, Wainberg MA. Genetic divergence of human immunodeficiency virus type 1 Ethiopian clade C reverse transcriptase (RT) and rapid development of resistance against nonnucleoside inhibitors of RT. Antimicrob Agents Chemother. 2002;46:2087–2094. doi: 10.1128/AAC.46.7.2087-2094.2002. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

