Polypurine-repeat-containing RNAs: a novel class of long non-coding RNA in mammalian cells
- PMID: 20940252
- PMCID: PMC4481618
- DOI: 10.1242/jcs.070466
Polypurine-repeat-containing RNAs: a novel class of long non-coding RNA in mammalian cells
Abstract
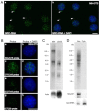
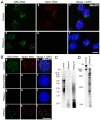
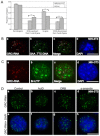
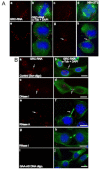
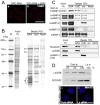
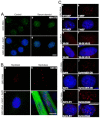
In higher eukaryotic cells, long non-protein-coding RNAs (lncRNAs) have been implicated in a wide array of cellular functions. Cell- or tissue-specific expression of lncRNA genes encoded in the mammalian genome is thought to contribute to the complex gene networks needed to regulate cellular function. Here, we have identified a novel species of polypurine triplet repeat-rich lncRNAs, designated as GAA repeat-containing RNAs (GRC-RNAs), that localize to numerous punctate foci in the mammalian interphase nuclei. GRC-RNAs consist of a heterogeneous population of RNAs, ranging in size from ~1.5 kb to ~4 kb and localize to subnuclear domains, several of which associate with GAA.TTC-repeat-containing genomic regions. GRC-RNAs are components of the nuclear matrix and interact with various nuclear matrix-associated proteins. In mitotic cells, GRC-RNAs form distinct cytoplasmic foci and, in telophase and G1 cells, localize to the midbody, a structure involved in accurate cell division. Differentiation of tissue culture cells leads to a decrease in the number of GRC-RNA nuclear foci, albeit with an increase in size as compared with proliferating cells. Conversely, the number of GRC-RNA foci increases during cellular transformation. We propose that nuclear GRC-RNAs represent a novel family of mammalian lncRNAs that might play crucial roles in the cell nucleus.
Figures
References
-
- Alexandre S., Rast C., Nguyen-Ba G., Vasseur P. (2000). Detection of apoptosis induced by topoisomerase inhibitors and serum deprivation in syrian hamster embryo cells. Exp. Cell Res. 255, 30-39. - PubMed
-
- Amaral P. P., Dinger M. E., Mercer T. R., Mattick J. S. (2008). The eukaryotic genome as an RNA machine. Science 319, 1787-1789. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

